

Institute of Bioanalytical Chemistry
Prof. Dr. Norbert Sträter
 |
 |
| Center
for Biotechnology and Biomedicine Institute of Bioanalytical Chemistry |
|
| Structural
analysis of
biopolymers Prof. Dr. Norbert Sträter |
|
Bacterial 5'-nucleotidases: Purine salvage and interference with purinergic signaling
|
|
Homologs of mammalian ectonucleotidases also exist in bacteria and other microorganisms, including many human pathogens. These enzymes have been demonstrated as virulence factors,
probably by either hydrolyzing extracellular nucleotides for purine
salvage or by generating extracellular adenosine in the host, which is
a powerful immunosuppressant signaling molecule. NTPDases as well as CD73 homologs have been found in many microrganisms.
First structural information on 5'-nucleotidase (5NT) became available by the crystal structure of E. coli 5NT, a zinc-containing enzyme. We characterized the structure of 5NT in open and closed conformations which differ in the relative orientation of the two domains by a rotation of up to 96° [1-4]. The domain movement can be described as a rotation of the C-terminal domain around an axis, which passes through the center of the C-terminal domain. The resulting hinge-bending domain movement is unique in that the cleft between the domains does not open up, but the residues of the domain interface slide along the interface. |
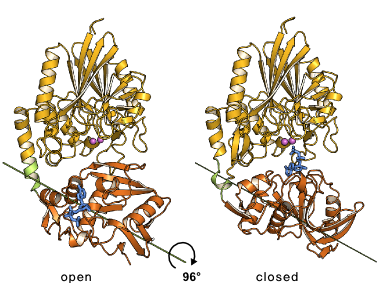 Domain rotation of E. coli 5NT
|
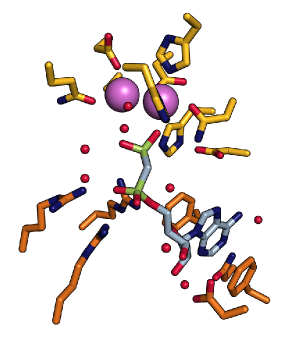 Active site of E. coli 5NT with the dimetal center and the substrate binding pocket |
Currently we study the relation between protein structure, function and the domain motion of 5NT employing especially NMR and EPR spectroscopy. We are also interested in the structure of 5'-nucleotidases from other human pathogens. |
|
Coworkers Thomas KnöfelRobert Schultz-Heienbrok Miriam Carl Susanne Moschütz Renato Weiße Matthias Zebisch Ulrike Krug Michel Krauß Troels E. Linnet Collaborations Professor Dr. Jochen Balbach,
Martin-Luther-Universität Halle-Wittenberg, Biophysik Publications
|