Mehr als 12 Jahre Transregio 67 Projekt: Funktionelle Biomaterialien zur Steuerung von Heilungsprozessen in Knochen- und Hautgewebe – vom Material zur Klinik
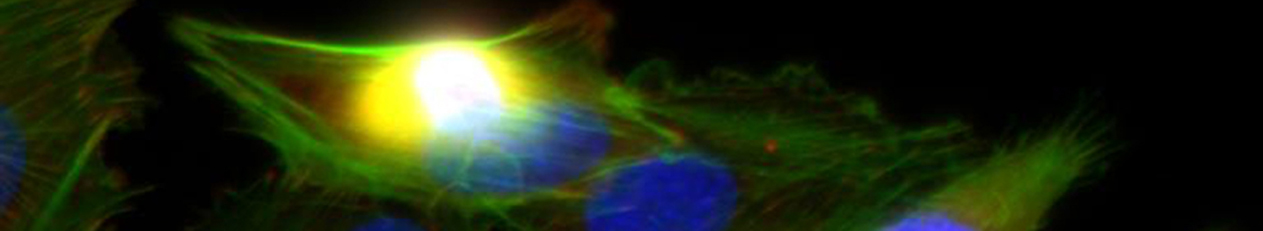
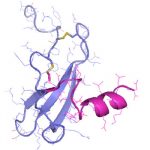
Die demographische Entwicklung in Deutschland und anderen Industrienationen bedingt eine deutliche Zunahme von Patienten mit Knochendefekten und chronischen Wunden bei gleichzeitig eingeschränkter Geweberegeneration. Dies stellt eine enorme klinische und sozioökonomische Herausforderung dar und erfordert die Entwicklung neuartiger funktioneller Biomaterialien zur Verbesserung der Knochen- und Hautregeneration in einer alternden, multimorbiden Bevölkerung. Dafür ist es besonders vielversprechend, das in den letzten Jahren gewonnene Wissen über die Bedeutung der extrazellulären Matrix (EZM) für die Regeneration von Geweben in die Entwicklung von Biomaterialien einzubeziehen. Die Struktur und Zusammensetzung der EZM beeinflusst entscheidend zelluläre Differenzierungsprozesse und Funktionen und damit die Heilung von Geweben. Vor diesem Hintergrund verfolgte der Transregio 67 (TRR67) das Ziel, neuartige funktionelle Biomaterialien auf der Basis artifizieller extrazellulärer Matrizes (aEZM) zu entwickeln und zu untersuchen. Wesentliche funktionelle Komponenten dieser Materialien waren Glykosaminoglykan (GAG)-Derivate und Proteoglykan (PG)-Analoga in Kombination mit Strukturproteinen oder synthetischen Trägersubstanzen.
Vision des Transregio 67 war, die gewonnenen Erkenntnisse über die Wechselwirkungen von GAG-Derivaten und PG-Biomimetika mit Mediatorproteinen, Matrixkomponenten und Zellen zu nutzen, um durch aEZM mit gezielt einstellbaren biologischen Eigenschaftsprofilen eine individuelle, an die Situation multimorbider Patienten angepasste Heilung von Knochen- und Hautgewebe zu erreichen.
Von 2009 – 2021 wurden in drei aufeinander aufbauenden Förderperioden grundlegende Erkenntnisse zu GAG/Mediator Wechselwirkungen gewonnen. Es wurde nachgewiesen, dass aEZM-basierte Biomaterialien Heilungsprozesse in Knochen und Haut fördern; die zugrundeliegenden Mechanismen konnten bis auf die atomare Ebene charakterisiert werden. Schließlich wurde die Komplexität der Biomaterialien erhöht, um sie an die in mehreren Phasen verlaufenden Regenerationsvorgänge in Knochen und Haut anzupassen.
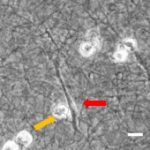
Diese multifunktionalen Biomaterialien wurden in einem translationalen Ansatz auf ihre Wirksamkeit in relevanten präklinischen Modellen mit eingeschränkter Regeneration in Knochen und Haut evaluiert.
Dieses Vorgehen bildete die Grundlage für die Entwicklung innovativer Therapiekonzepte zur Behandlung chronischer Wunden und mündete mit Ende der letzten Förderperiode in die Phase I einer klinischen Prüfung am Menschen.
Der TRR67 war vom 1. Juli 2009 – 31. Dezember 2021 ein gemeinsames Projekt der Universität Leipzig und der Technischen Universität Dresden unter Beteiligung der Freien Universität Berlin. Als außeruniversitäre Forschungseinrichtungen waren das Helmholtz-Zentrum für Umweltforschung Leipzig-Halle GmbH, das Helmholtz-Zentrum Dresden-Rossendorf, das Leibniz-Institut für Polymerforschung Dresden e.V., INNOVENT e. V. und als Industriepartner die Firma Mathys Orthopädie GmbH beteiligt.
Er gliederte sich in vier Projektbereiche:
- Schwerpunkt im Projektbereich „Matrixengineering“ war die Erforschung neuer GAG- und PG-Derivate, aEZM, Trägersubstanzen und Beschichtungen in ihrer Wechselwirkung mit Schlüsselmediatoren der Knochen- und Hautregeneration.
- Im Projektbereich „Biologische Wirkprofile“ wurden die neuen Biomaterialien in für die Knochen- und Hautregeneration relevanten Zellkulturen, 3D-Kultursystemen und therapierelevanten präklinischen Tiermodellen funktionell charakterisiert.
- Im Projektereich „Transfer“ wurden unter Beteiligung industrieller Partner die Voraussetzungen für eine spätere klinische Erprobung der neuen Biomaterialien geschaffen.
- Der „zentrale Zuständigkeitsbereich“ umfasste Serviceprojekte, die Verwaltung und das Integrierte Graduiertenkolleg (IGK) zur strukturierten Promovierendenausbildung.
Besonders hervorzuheben am TRR67 war sein von Anfang an interdisziplinärer Ansatz mit Beteiligung von Wissenschaftler:innen aus Materialwissenschaft, Bioinformatik, Biophysik, Biochemie, Glyko- und Medizinischer Chemie, Pharmazie, Zell- und Matrixbiologie, Immunologie und klinischer Medizin sowie die intensive Kooperationskultur, die auch über die Förderdauer des TRR67 hinaus fortbesteht.
Ansprechpartner

Sprecher des TRR 67
Prof. Dr. Jan-Christoph Simon
+49 (0)341 97-18600
jan.simon@medizin.uni-leipzig.de

Stellvertretender Sprecher
Prof. Dr. Carsten Werner
+49 (0)351 4658 531
werner@ipfdd.de
News & Events
Da das Forschungsprojekt abgeschlossen ist, wird die Website ab 1. April 2022 nicht mehr aktualisiert.
TRR67 Highlight Issue – Extracellular Matrix Engineering for Advanced Therapies
Video Abschluss-Retreat – Mehr als 12 Jahre TRR67 Projekt
TRR67 Symposium
3rd INTERNATIONAL SYMPOSIUM
of the Transregio 67
in Leipzig
News & Events
Auf dieser Seite finden Sie eine Zusammenstellung aktueller Nachrichten und Veranstaltungen aus unserem TRR 67.
TRR67 Seminare
Die aktuellen Seminare im Rahmen der Graduiertenausbildung sind auf dieser Seite zu finden.
Neue Publikationen
Im Rahmen des Transregio sind zahlreiche Publikationen entstanden, die Sie im folgenden finden.