PyMOL Tutorials (using the GUI): Overview of the protein fold
A typical representation of a protein structure shows the complete fold
together with residues important for function (receptor binding pocket,
active site). Of course, the way you approach this task depends on your
knowledge about the structure and function of the protein as well as about
functionally important residues and ligands. For the current tutorial, we
assume that we know next to nothing about the protein.
Menu: File, Reinitialize, Everything: This resets PyMOL to a state
as if the applications was just started with all default settings.
Menu: File, Get PDB... 4eiy
To be completely independent of any default settings, the current PyMOL
version might provide, we start from scratch and hide all automatically
generated representations:
OB: all: "H" Hide, everything
We first show the fold of the protein as cartoon: OB: 4eiy: "S" Show,
cartoon. Rotate the structure and inspect the helical fold. You may
notice that some loops are represented as if they were helical regions. We
therefore choose OB: 4eiy: "A" Action, assign sec. struct. to
improve the quality of secondary structure assignment.
Figures used for publication usually have a white background: Menu:
Display, Background, white.

|
A rather special problem with residue numbering of the BRIL
insertion domain
Have a closer look at the connectivity of the fold. You may notice
that there are four α-helices beneath the 7TM domain that appear to
consist of 2 x 2 separated helices. One reason for this phenomenon
might be disordered loops, which are not resolved in the crystal
structure. These can be represented by broken lines. Choose Menu:
Setting, Cartoon, Gap Cutoff, 20. Gaps in the protein sequence
are connect be a broken line, if the distance is shorter than the
cutoff value in Angstrom, which we just increased from the default
value of 10 Å to 20 Å. Now two helices are connected correctly by
the lines indicating a disordered loop, but two transmembrane
helices are also connected and they appear to be disconnected from
the 4-helix domain.
The reason for this strange behavior is that the construct used
for crystallization contains a BRIL domain (a small protein) fused
into its third intracellular loop (ICL3) to facilitate
crystallization of the construct. The reason for the breaks is the non-continuous
residue numbering at the fusion sites. We can analyze this by
clicking at the residues at the gaps. Just click on the cartoon
ends flanking the gaps with the left mouse button. This
selects the atoms of the residue and a selection string of the
clicked atom is displayed in the Output window. At one of the gaps
the adjacent residues are listed as:
"You clicked /4eiy/A/A/GLU`219/CA" and
"You clicked /4eiy/A/A/LEU`1106/CA".
These selection macros have the
format /object/seg-id/chain-id/resname`resid/name, where 4eiy is the
name of the object, A is the segment identifier, A is the chain id,
GLU is the residue name, 219 is the residue number, and CA is the Cɑ-atom.
Notice that there is a large gap (difference) between the residue
numbers 1106 and the following residue number 219 in the sequence.
The residue numbers of the BRIL domain have been incremented by 1000
relative to the "correct" numbers (obtained from the protein
sequence position) to avoid that the fusion insertion generates
duplicate residue numbers in the same chain.
|
Unfortunately, there is no simple solution to show the cartoon fold in a
continuous representation. One could probably renumber the residues of the
BRIL domain such that they fall in the range of ICL3 residues replaced by
this domain. With the help of insertion identifiers, several residues can
share the same residue number. As this is a tedious modification, unless
we write a little script that does this job, we will prepare two
representations of the GPCR fold. In one figure we show the BRIL
domain and connect it via a line to the 7TM domain. In the second image,
we simply omit the BRIL domain.

|
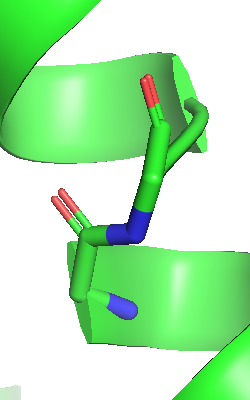
To bridge the gap between the 7TM and
BRIL domains, click with the left mouse button on the cartoon at one
side of one of the gaps and next with SHIFT-left-mouse click on the
cartoon edge at the other side of the gap. This will select all
atoms of the two residues at the interface between 7TM and BRIL
domains. The selected atoms are shown as pink dots as in the image
shown here. In the object menu, a new menu line named "(sele)" has
appeared. The brackets indicate, that this is not an object, but a
selection. This selection named "sele" now enables us to represent
the selected atoms via the OB: (sele) S: Show options. As we
only want to bridge the gap, we show the main chain atoms via OB:
(sele) S: Show: main chain, sticks. The main chain atoms of
the linkage are now shown. |
|
|
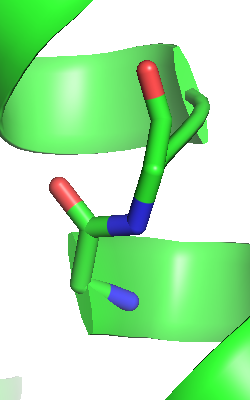
The default representation of sticks in
PyMOL is such that double bonds are shown as two smaller bonds which
facilitates the recognition of chemical structures. While this may
be useful for bound ligands, the classical representation of bonds
as a single sticks is nicer at least for protein residues in an
overview cartoon fold figure. You can toggle between the two
representation modes of sticks via the Menu: Display, Show
Valences option.
Next, connect the two ends of the other gap in the same way.
After that, we reduce the cartoon gap cutoff to 10 Angstrom again,
as wrong termini are connected in this special example: Menu:
Setting, Cartoon, Gap Cutoff, 10. |
|
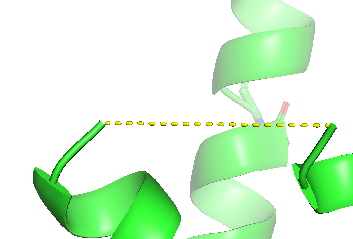
To represent the connection of the long
disordered loop of the BRIL domain, the ends may be connected by a
straight broken line. This is achieved via the GUI by the
Measurement Wizard: Menu: Wizard, Measurement. Click at the
two ends of the gap. The distance is represented by a broken line
and the distance is indicated. Select "Done" in the new Measurement
menu of the object menu. We remove the distance label by OB:
measure01: H: Hide, labels. |
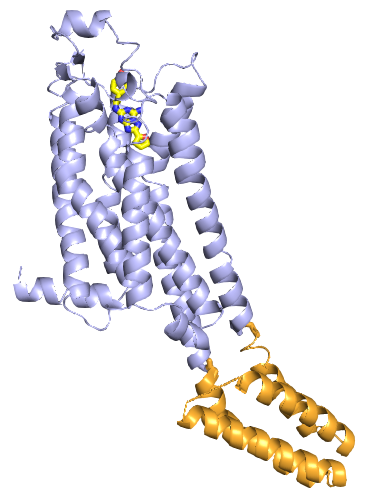
Next, we want to represent the 7TM and BRIL domains in different colors.
As far as I know, there is no option to select a range of residues via the
mouse by clicking on the residues directly. Instead, we display the
sequence via Menu: Display, Sequence.

The sequence is colored in green as is the fold. The sequence parts in
grey are disordered residues which are listed in the pdb file but
do not have coordinates in the structure. The residues starting with 1001
are the BRIL domain. Select the first residue A1001 by left mouse click on
the sequence position and next click on the last residue via
SHIFT-left-mouse on L1103. The residues of the BRIL domain are selected in
the sequence and structure. Choose OB: (sele) C: Color: oranges,
brightorange. To deselect all atoms, click somewhere where there is
no atom. Next select the 7TM domain and color it in lightblue. Color the
broken line of the BRIL domain (measure01) also in brightorange.
|
In addition to the fold, we also want to
show the location of ligand binding. We therefore select
the ligand residue ZMA via the sequence. Obviously, we need to
know the ligand residue name to do this. You may have this
information for example from the pdb entry of the 4eiy structure in
the RCSB protein data bank, which shows chemical structures of the
ligands. Alternatively, you display all atoms via OB: 4eiy S:
Show: lines to locate the ligand and left-click on the ligand
to obtain the residue name and number.
We want to have the ligand generated as a separate object. To this
end, after selecting the ligand, use OB: (sele) A, Action: copy
to object, new. A new object with name "new" is generated. We
change the object name to "ligand" via OB: obj01 A, Action:
rename object. Enter in the Display Area the new object name
"ligand". Next, display the ligand as sticks via OB: ligand, S
Show: sticks. Change the atom colors via OB: ligand, C:
Color: by element, CHNOS with yellow carbon atoms. Orient the
molecule in a favorable view. You may notice that the ligand is not
well visible at large distance. To improve this, the stick radius
may be increased. Unfortunately, Menu: Setting, Lines &
Sticks, Stick Radius provides no values larger than the
default 0.25. We therefore have to use the Command Line,
detailed later: "set stick_radius, 0.6". |
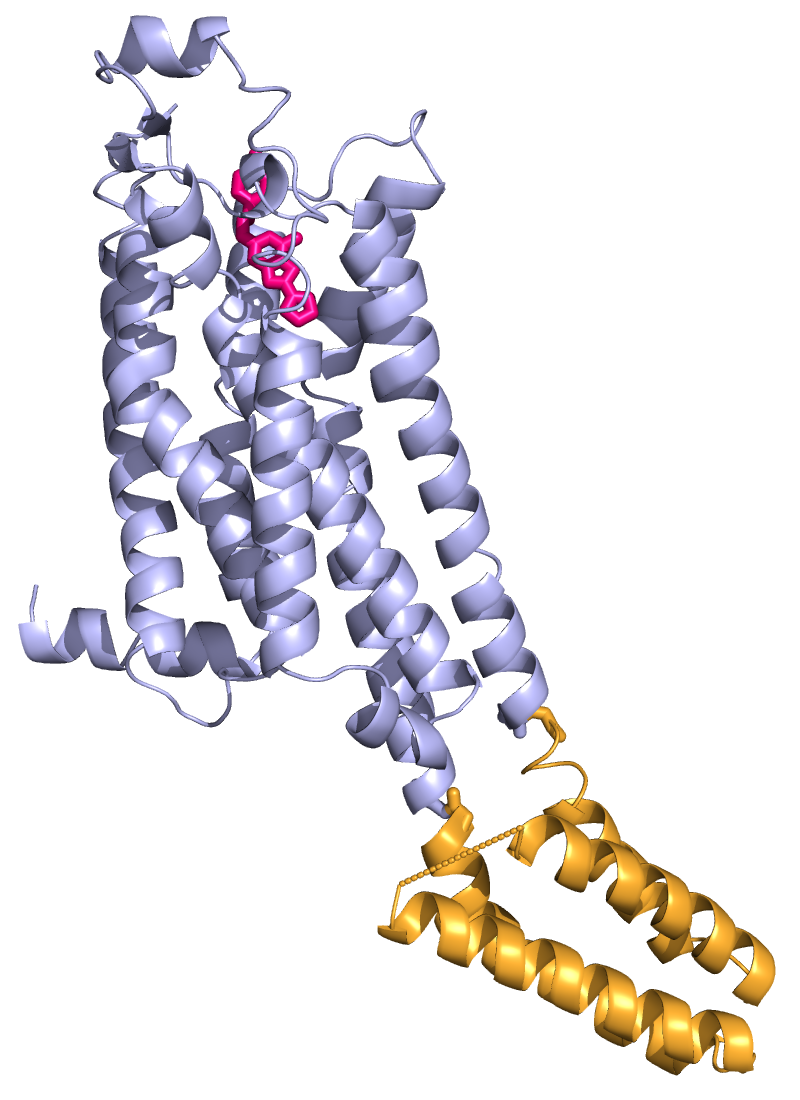
|
To further increase the visibility of
ligands at large distance, it is appropriate to use a single
color for all atoms and to use colors that provide good
contrast from the fold. In the example shown here the ligand has
been colored by OB: ligand: C: Color: magentas, hotpink.
To optimize the quality of the image by changing a number of preset
parameters, use: Menu: Display, Quality, Maximum Quality.
To raytrace the image and generate it an appropriate
resolution and aspect ratio, zoom the molecule to fit (usually)
the vertical dimension. Next play around with the draw
command and the aspect ratio. The resolution you need depends
on the final size in which the image will be displayed. For a print,
a one column figure should be ~1000 pixels in width. To generate the
figure you use the command draw x_pixels, y_pixels in the
Command Line. When you are finished, you may use the ray (ray
tracing) command for the final figure. The image shown here
was generated with ray 800,1100. Usually, you also need to center the
molecule via the middle mouse button (shift it in the image
plane).
To save the image use the command: png my_filename.png.
Before you do this, change to a directory, where you want to
store the image: cd d:\my_projects\my_directory.
|
Transparent background: I have generated the image with a white
opaque (non-transparent) background. The default is a transparent
background. To make the background opaque, use the option Menu:
Display, Background, Opaque. If you ray-trace with transparent
background, a chessboard pattern is displayed as background to indicate
the transparency. You can change (toggle) the chessboard pattern to a
white background via Menu: Display, Background, Alpha Checker. The alpha channel is a
color component that represents the degree of transparency (or
opacity) of a color. Transparent background is preferable if you need
to arrange several images close to another in a composite image and if
you want to avoid that the opaque background is ontop of a neighboring
image. Or if you show the fold ontop of another image as background.
If you do not need this, it is preferable not to use transparency as
it can cause trouble in certain situations.
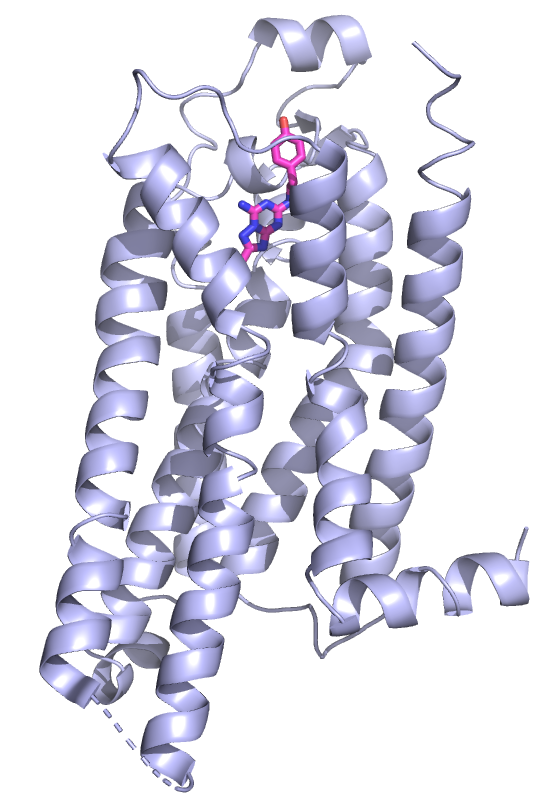
|
In a second image we want to omit the
artificial BRIL domain and focus on the 7TM part of the receptor. To
omit the BRIL domain, select the corresponding residues via the
sequence view and hide the cartoon via: OB: (sele) H, Hide:
Cartoon.
In the image shown here I colored the ligand with pink carbons and
used a stick_radius 0f 0.4. The gap_cutoff was increased to 20
Angstroms to indicate the disordered cytosolic loop (Menu:
Setting, Cartoon, Gap Cutoff, 20). This works now fine as the
BRIL domain is not shown.
Next we use some alternative representation of the complete fold
together with the ligand. One frequent option is to color domains or
subunits differently, but as the 7TM domain is a single domain, this
is not of interest here.
|

|
In the image shown to the left, the fold is
colored from N-terminus (blue) to C-terminus (red) in rainbow
colors. If you apply OB: 4eiy, C: Color spectrum, rainbow(*/CA),
the rainbow ends somewhere in yellow-orange colors as the not shown
BRIL domain with its high residue numbers is included. Therefore,
you mark only the 7TM domain via the sequence and apply OB:
(sele), C: Color spectrum, rainbow(*/CA) to obtain the
desired coloring.
In the image shown to the right, two further settings were applied:
Menu: Setting, Cartoon, Fancy Helices and Menu: Setting,
Cartoon, Highlight Color. The first option results in the
smoother shape of the helices at their ends. This option is
generally recommended. The second option colors the inside of the
helices in a different color, default is the grey color shown here.
You can change this color, i.e. set cartoon_highlight_color,
bluewhite. Check the OB menus for available color names.
|
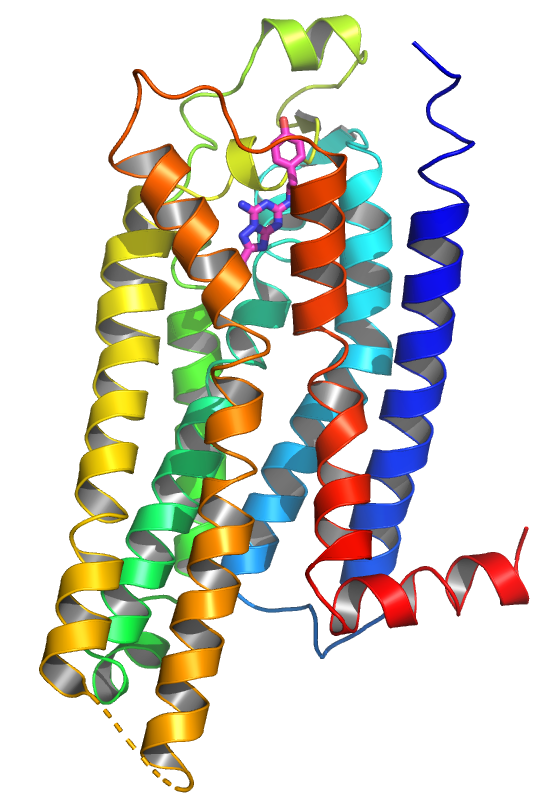
|

|
Adding labels. In a scientific paper
you usually add text to the images to provide more information. This
feature is provided by the label command in PyMol. But you have to
options. You either add the labels in PyMol or you add them later in
inkscape. I prefer the latter option, as text editing is much more
versatile in inkscape. But there may be situations, where you prefer
to add labels in PyMol, so we add labels to the current image.
Assume we want to number the transmembrane helices. I think there is
no option to directly add an arbitrary text label by clicking on a
residue and using the right-mouse context menu. So we need to use a
command in the Command Line. The general syntax to use is:
label <selection>, "your_text"
We will discuss selection syntax later. For now, the easiest
option is to click on a residue somewhere in the middle of the blue
helix, which is transmembrane helix 1 (TM1). In the Output window a
selection string of the form "/4eiy/A/A/LEU`19/CA" is returned, as
already mentioned above. By copy/paste from the Output window we use
this string in the Command line to place our label:
label /4eiy/A/A/LEU`19/CA, "TM1"
Add labels to all seven transmembrane helices in this manner. You
will notice that the labels are somewhat small and some may be
hidden partially by the cartoon. Using the command Menu:
Setting, Label, Size, 24 Point we increase the label size.
With the command set label_position,
(0,0,10) we move the
labels by 10 Angstrom towards the viewer, such that they are not
hidden by any cartoon parts. Next, you may want to shift the
labels in the x and y directions (in the plane) to a favorable
position. Apart from using the above command set label_position, (x,y,z), we can also do this interactively by
clicking on the line "Mouse Mode 3-Button Viewing" in
the Mouse controls window (lower right of GUI). This will
activate the "3-Button Editing" mode and we can now cntrl-left-click
on the label to move it (keeping ctrl pressed). Be
careful not to miss a label and hit an atom instead, as this
will move the (I guess) C-alpha atom in this case and
distort the structure. When you are finished, toggle back to
3-Button
Viewing Mouse Mode by clicking on the Mouse controls
window again.
In the figure shown to the left, I also used Menu:
Setting, Label, Background Color, Back, which places
the semitransparent boxes behind the labels to increase its
visibility against the dark fold.
|
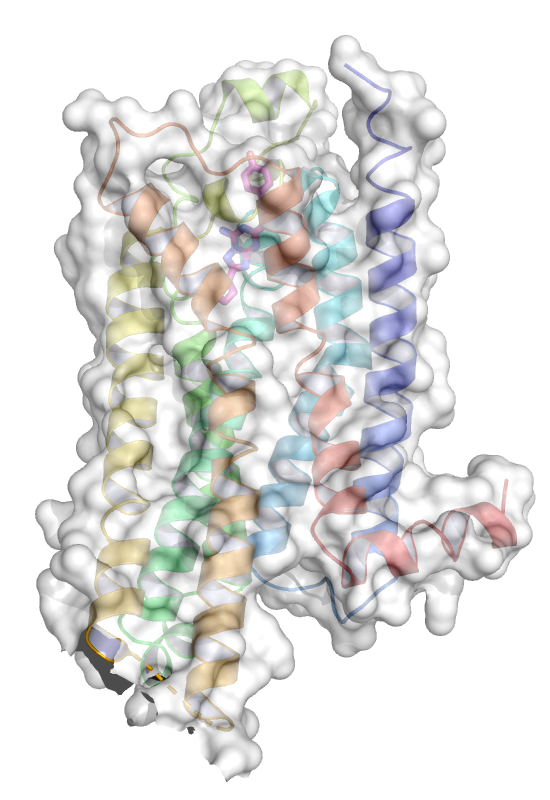
|
Another common option is to show the molecular
surface. The figure to the left shows the molecular surface of
the 7TM domain. The following options were used to generate this
image starting from the previous representation.
In the Command line: hide labels. (Alternatively hide labels
using the OB menus). Next, the molecular surface is generated by
selecting the 7TM domain in the sequence window and OB: (sele) S
Show: surface. Using Menu: Setting, Surface, Color, White
and Menu: Setting, Transparency, Surface, 40% the surface is
shown in transparent white.
Showing the molecular surface is not a very useful representation in
this case. It is more frequently used to
- show large assemblies represented by its parts (domains,
subunits, proteins) in different colors
- show the fit of a ligand to its binding site
- map properties to the surface, e.g. electrostatic potential or
conservation
|
Back to PyMOL tutorial main page.












