(1) Add low-energy hydrogen positions to the structure using the ProteinsPlus webserver.
(2) To show hydrogen bonds within a ligand and its environment use one of the following procedures.
| (A) Show H bonds without hydrogens |
(B) Show H bonds with hydrogens |
| reinitialize load 4eiy_protoss.pdb hide all select ligand, resn ZMA select env, byres (ligand around 5.0) and not ligand show sticks, ligand or env show nb_spheres, env set h_bond_max_angle, 40 set h_bond_from_proton, 0 dist hbonds, ligand or env, ligand or env, mode=2 remove hydrogens set dash_gap, 0.4 set dash_radius, 0.05 set dash_length, 0.3 hide labels, hbonds color grey50, hbonds color gold, ele C and ligand color bluewhite, ele C and env orient ligand or env |
reinitialize load 4eiy_protoss.pdb hide all select ligand, resn ZMA select env, byres (ligand around 5.0) and not resn ligand show sticks, ligand or env set h_bond_max_angle, 40 set h_bond_from_proton, 1 dist hbonds, ligand or env, ligand or env, mode=2 set dash_gap, 0.4 set dash_radius, 0.05 set dash_length, 0.3 hide labels, hbonds color grey50, hbonds color gold, ele C and ligand color bluewhite, ele C and env orient ligand or env |
For further details and other options, see below.
dist obj_name, selection1, selection2, mode=2
Mode 2 enables the detection of hydrogen bonding interactions based on distance and angular criteria. However, there are two main points to consider. First of all, angular geometry criteria can only be used if the positions of the polar hydrogens are known. We discuss this in a following section.
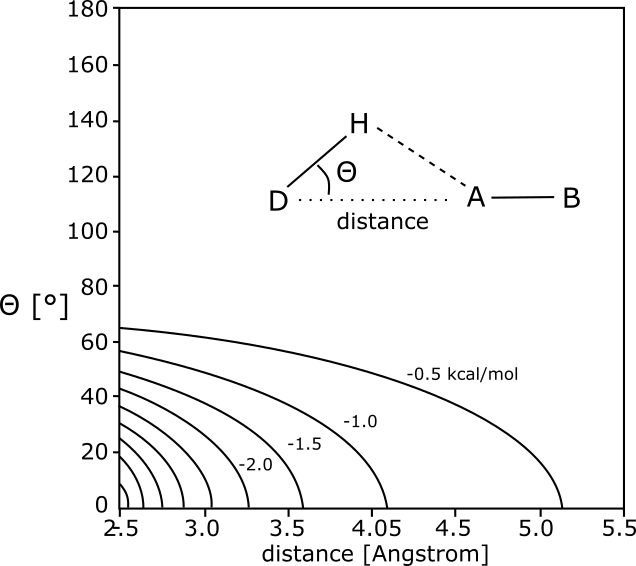
For determination of the hydrogen bonds via the dist command with mode=2, PyMOL follows a concept described by Kabsch & Sander, Biopolymers 22, 2577 (1983). The following figure schematically illustrates this strategy.

| parameter |
default |
meaning |
| h_bond_cone | 180 | The present value (180°) rejects any hydrogen bond where the hydrogen would lie behind the plane defined by the acceptor atom (A) in relation to its bonded atom(s) B (if any). For example, a D-H...O=C hydrogen bond will be rejected if the H-O-C angle is less than 90°. I think, this parameter should not be changed. |
| h_bond_cutoff_center | 3.6 | This is the maximum distance for ideal hydrogen bonds. |
| h_bond_cutoff_edge | 3.2 | This is the maximum distance for a marginal hydrogen bond. |
| h_bond_max_angle | 63.0 | Defines the maximum angle Θ for a marginal hydrogen bond (see figure above for the definition of the angle Θ). This value appears to correspond to the maximum misalignment of 63° allowed at ideal length (2.9 Å) in the Kabsch & Sander paper. |
| h_bond_power_a | 1.6 | |
| h_bond_power_b | 5.0 | The two h_bond_power_* settings are described as fitting parameters to reproduce the curve shape of the Kabsch & Sander paper. The endpoints of the effective cutoff curve are a function of the two h_bond_cutoff_* setting. |
| h_bond_from_proton | 1 | This controls if the hydrogen bonds are drawn between the hydrogen atoms and the acceptor (1) or between the donor and and acceptor atoms (0). The hydrogen bonds will only be drawn from the hydrogen to the acceptor, if a hydrogen atom is present and it will be drawn between the heavy atoms, if not. |
It is not obvious, how these parameters generate the energy function of the above figure in detail. Also the absolute and relative magnitudes of the two h_bond_cutoff_* parameters are not clear to me. One would probably have to study the PyMOL python code of these functions, to understand this. However, after analyzing the hydrogen bonds drawn for a structure with reasonable hydrogen positions, it appears as if the very generous distance criteria of the Kabsch & Sander publication (Figure 1, allowing for distances up to 5.2 Ang.) have been reduced to reasonable values, whereas the angle cutoff parameter has a generous default value of 63°. Therefore, you may want to reduce h_bond_max_angle to values such as 40 to show only significant hydrogen bonding interactions in agreement with the chosen distance settings.
However, be aware that you cannot fully trust this command and you have to check the interactions. I noticed a number of cases where the donor and acceptor heavy atoms are connected instead of the hydrogen atom or where no hydrogen bonding interaction is detected despite perfect distance and donor angle geometry.
For most experimental structures, hydrogen atom positions are not defined experimentally, although they may be present in the pdb file for some atoms to improve refinement. Some hydrogen positions (e.g. amide hydrogens) can be added accurately based on the heavy atom coordinates, but for alcohol groups or water molecules, this is not possible. If ligands are present in the structure, their chemical structure (bond order, hybridization, ...) most be defined or deduced from the atomic coordinates. This information is not part of the pdb or cif files describing the protein structures. Determination of reasonable low-energy positions of all hydrogen atoms is a non-trivial task, but this can be achieved by professional software dedicated to molecular modelling.
PyMOL can add hydrogen positions via the h_add command. However, the positions of hydrogens that are not fixed by the positions of the heavy atoms will not be placed to achieve favorable interactions with its environment. Therefore, this is not the recommended option.
A free and good choice is the software PROTOSS which can be used via the ProteinsPlus webserver. You can upload a pdb file or access structures in the PDB directly via the identifier. Next you choose the Protoss hydrogen prediction software and hit the red buttons "Protoss" and next "Calculate". Via the "Download PDB button" you save the pdb file with the hydrogen coordinates to your hard disk. Name the file *_protoss.pdb, so that you remember that this is a file from Protoss.
If you have the commercial software MOE installed, you can read the file into MOE and use the options "Compute, Prepare, Structure Preparation" and next "Compute, Prepare, Protonate3D". After that, save the file with the hydrogen positions as *_protonate3D.pdb.
Strategy 1: Add hydrogens, determine hydrogen bonds using angle criteria, add missing hydrogen bonds manually
(1) add hydrogens using protoss or other suitable software
(2) determine H-bonds using the commands above (usually without hydrogens shown)
(3) check the environment: omit residues which shall not be shown and check if hydrogens bonds are missing
(4) add missing hydrogen bonds using the dist command and selection macros (you get these from clicking on the atoms)
Strategy 2: Specify all hydrogen bonds manually via dist commands
A tedious but very flexible option is to add the hydrogen bonds manually one after the other using this dist command with two selection macros.
Use the PoseView program of the ProteinsPlus webserver to determine hydrogen bonds
independently for comparison.
Link to tutorial:
Generating ligand interaction images
Back to PyMOL tutorial main page.