DNA-binding functions of PepA
Biological background
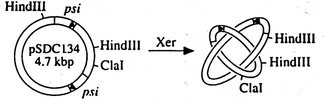
E. coli PepA acts as a assessory protein in Xer site specific
DNA recombination. Plasmid multimers are formed in E. coli by
homologous
recombination. These plasmid multimers reduce the number of independent
plasmids and increase the chance of forming plasmid-free daughter cells
at cell division. The Xer system resolves plasmid multimers, but does
not
form new multimers, i.e. the recombination reaction is exclusively
intramolecular.
For this selectivity the assessory proteins PepA and the arginine
repressor
(ArgR) are involved in addition to the two recombinases XerC and XerD.
In addition to this
function, PepA has an independent role in the transcriptional
regulation of the carAB operon, which encodes the enzyme
carbamoylphosphate
synthetase. For both functions interaction of PepA with DNA has been
demonstrated.
DNA-binding groove of PepA
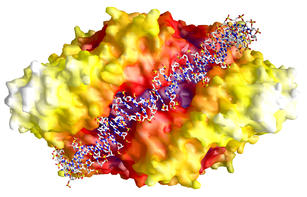 |
The molecular surface of PepA reveals a groove
which runs from
one trimer face along the two-fold molecular axis to the other trimer
face.
The model shown here demonstrates that this groove is large enough to
accomodate
duplex DNA.
When the DNA winds around PepA in the direction of the three-fold axis
(from top to bottom in this figure), the topology of the hexamer
directs
the DNA strand to form a right-handed superhelix. This is in agreement
with the formation of right-handed catenanes as plasmid products. |
A model for the Xer complex
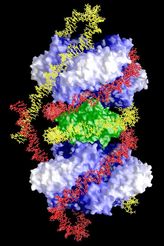 |
Based mainly on the topology of the plasmid
products (4-noded,
right-handed catenanes), DNase I footprinting assays and the X-ray
structures
of PepA and ArgR, a new model for the Xer complex was proposed by us,
in
which two PepA molecules (blue) and one arginine repressor (green)
associate
along the trimer faces. The two recombination sequences (red and
yellow) are wound around the protein complex such that right-handed
superhelices
are formed and three nodes are trapped. An additional node is
introduced
in the recombination reaction.
Models were build for these complexes in order to have a realistic
estimation of the relative positions of the protein binding sites along
the DNA sequence and to ensure that the DNA curvature has realistic
values. |
References
Sträter, N., Sherratt, D. J. & Colloms,
S. D. (1999). X-ray structure of aminopeptidase A from E. coli and a
model
for the nucleoprotein complex in Xer site-specific recombination. EMBO
J. 18, 4513-4522.
Colloms, S. D., Bath, J. & Sherratt, D. J.
(1997). Topological selectivity in Xer site-specific recombination. Cell
88,
855-864.
|



