Mechanisms of Protein Kinase C (PKC) Translocation
back
Protein kinases C (PKC) are members of the AGC-kinase superfamily of serine/threonine protein kinases. The 10 known PKC isoenzymes are grouped into classical, novel and atypical isoenzymes based on their regulatory properties and domainx structure. Originally discovered by Yasutomi Nishizuka as Ca2+- and phospholipid-dependent protein kinases, members of the PKC family as well as PKC-related kinases (PRK´s) are present in virtually all cells of our body and regulate a vast array of cellular responses. Since most cells co-express at least one classical and one novel PKC, the question emerges, how signalling is maintained. The common catalytic core domain of Protein kinases is controlled by one ore more N-terminally positioned regulatory domains which comprise a diacylglycerol-binding, cysteine repeat-containing C1 domain (classical and novel PKCs) and a Ca2+ ion-binding C2 domain (only fully functional in classical PKCs). These regulatory domains are likely to add specificity to the complex network of PKC signalling.
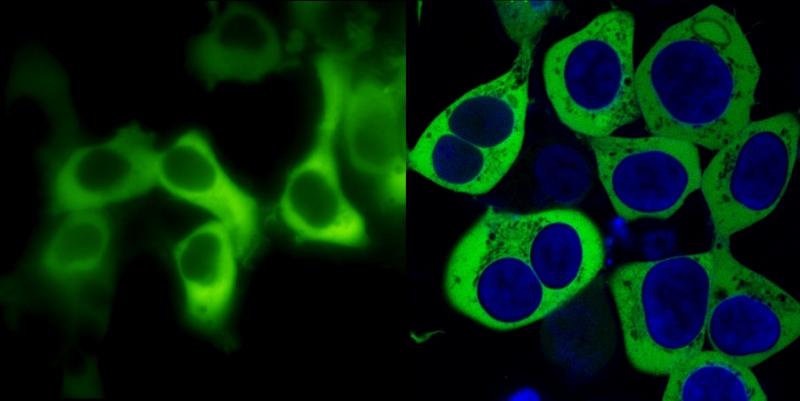
Fig. 1: PKCa C-terminally fused to GFP and imaged in living cells
The cDNA of human PKCa was amplified by PCR, the stop codon was replaced by an EcoR-V restriction site and in-frame ligated to the open reading frame of enhanced GFP in a mammalian expression vector pcDNA3-GFP. Upon transient transfection in HEK293 fibroblasts, fluorescence was imaged in living cells either in a conventional monochromator- and CCD-camera-equipped digital video imaging system (left panel) or in a confocal laser scanning microscope (right panel; nuclear DNA counterstained with the blue fluorescent dye bisbenzimide). Confocal pinholes were set to yield an optical slice of 0.8 µm.
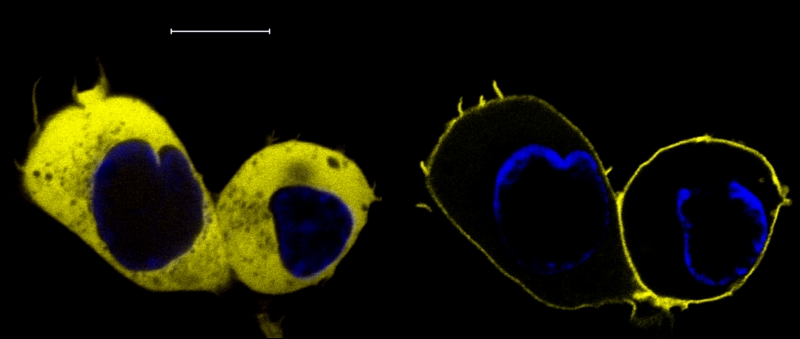
Fig. 2: Receptor-induced translocation of PKCa-YFP
PKCa-YFP
was imaged by confocal microscopy before (left panel) and 3 s after
stimulation of a coexpressed H1 histamine receptor with 100
µM histamine. Bar: 10 µm.
The receptor-induced and Ca2+-driven
translocation of classical PKC isoenzymes proceeded with a half-maximal
about 400-700 ms after agonist stimulation as assessed by
simultaneously recording the [Ca2+]i
and PKC translocation in fura-2 loaded cells. Considering that a mean
lateral movement of about 4-6 µm is required in order to
reach the plasma membrane, a diffusion-driven random movement of PKC
would result in 2-3 collisions with the plasma membrane per second.
Hence, in order to explain the fast kinetics of classical PKCs, an
unusually high collisional coupling efficiency at the plasma membrane
would be required.
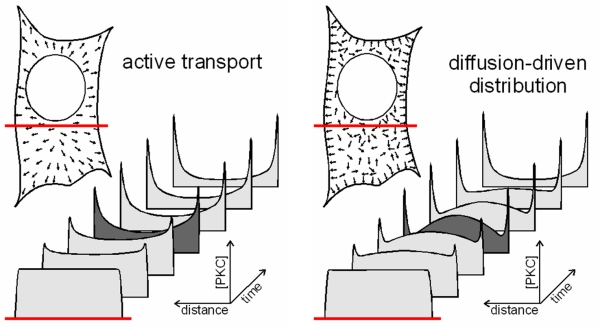
Fig. 3: Directional and random-oriented movement patterns result is a distinct subcellular distribution during the ongoing translocation process
An active transport towards the plasma membrane would be characterized by a directional movement and primarily deplete the inner parts of the cell. By contrast, a diffusion-driven and diffusion-limited translocation would primarily deplete the sub-plasmalemmal area until additional PKC molecules diffuse to the periphery of the cell. In the latter case, the global translocation speed would be largely limited by the velocity of PKCs diffusing towards the plasma membrane.
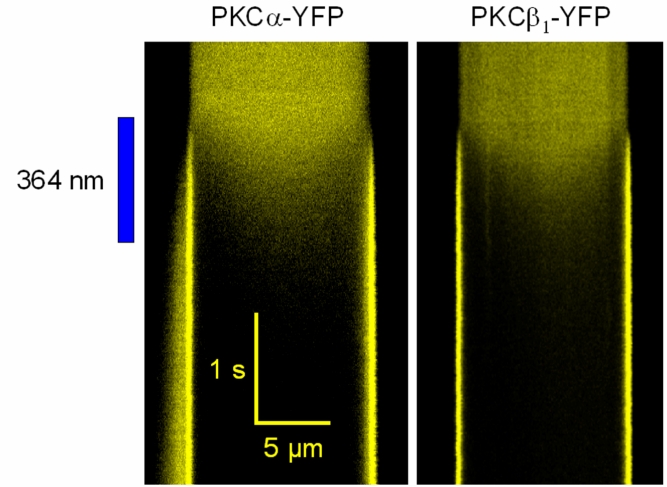
Fig. 4: Highspeed confocal line-scan imaging reveals a diffusion-limited association kinetics
PKCa-YFP (left panel) or PKCb1-YFP (right panel) were imaged in the cytosol of living cells. The horizontal dimension corresponds to an intensity profile along a line defined through the plasma membranes and the cytosol of living HEK293 cells that expressed the respective PKC fusion protein. The time preceeds from the top to the bottom of the image as indicated by the bars. PKC translocation was induced by applying a high local intensity of 364 nm light to uncage intracellularly loaded Ca2+ (cells were preincubated for 20 min with 5 µM o-nitrophenyl-EGTA). Note that during the ongoing membrane association of classical PKCs, a local concentration minimum occurs just below the plasma membrane. Since the inner part of the cells are being depleted only with a detectable delay, the translocation patern is indicative of a diffusion-driven and diffusion-limited translocation of classical PKCs.
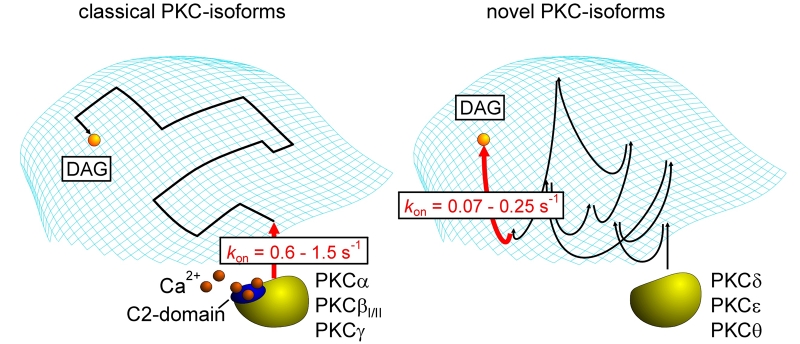
Fig. 5: Plasma membrane translocation and diacylglycerol binding of classical and novel PKC isoenzymes
Schematic drawing illustrating the distinct modes of trafficking of classical (left panel) and novel (right panel) PKC-isoforms. Upon receptor stimulation, clasical PKC isoenzymes attach to the plasma membrane in a diffusion-limited Ca2+- and phosphatidylserine-dependent mode and require only few collisions with the plasma membrane to undergo an electrostatically assisted and highly cooperative binding event. The subsequent scanning for diacylglycerols (DAG) is restricted to the two-dimensional membrane compartment. By contrast, novel PKC isoforms are lacking a functional Ca2+-binding C2 domain and, therefore, have to undergo multiple collisions at the plasma membrane until they eventually hit a diacylglycerol molecule. Typical kon values for the imaging-based assessment of the PKC membrane association are given.
back
Original articles of the group related to PKC translocation:
Schaefer M, Mischak H, Schnell S, Griese A, Iakubov R,
Riepenhausen G, Schöfl C (2004)
Mechanisms
of arginine-vasopressin-induced Ca2+
oscillations in beta-cells (HIT-T15): a role for oscillating protein
kinase C. Endocrinology 145:4635-4644.
Sinnecker D, Schaefer M (2004)
Real-time
analysis of phospholipase C activity during different patterns of
receptor-induced Ca2+ responses in HEK293 cells.
Cell Calcium 35:29-38.
Lenz JC, Reusch HP, Albrecht N, Schultz G, Schaefer M
(2002)
Ca2+-controlled
competitive diacylglycerol binding of protein kinase C isoenzymes in
living cells. J. Cell Biol. 159:291-302.
Schaefer M, Albrecht N, Hofmann T, Gudermann T, Schultz
G (2001)
Diffusion-limited
translocation mechanism of protein kinase C isotypes. FASEB
J. 15:1634-1636.