

Y2R_active_complex.pml
Y2R_active_complex_surface.pml
set transparency, 0.5, Y2
For this tutorial we choose the CryoEM structure of the Y2 receptor in complex with a peptidic ligand and heterotrimeric G protein, deposited as pdb id 7yon. From the pdb website (www.rcsb.org/structure/7YON) we obtain information on the protein chains and ligands in the complex, summarized here:
| chain |
molecule |
| A | Guanine nucleotide-binding protein G(i) subunit alpha-1 |
| B | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
| G | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
| L | Peptide YY(3-36) |
| R | Neuropeptide Y receptor type 2 |
| S | single-chain antibody Fv fragment (scFv16) |
Tasks:
Generate an overview figure that shows all protein chains of the complex
in different colors, but omit the "unphysiological" antibody fragment,
that has been added to facilitate CryoEM analysis. In addition to the
cartoon representation, the complex shall be represented via the molecular
surfaces and a combination of cartoon and surface.
Write a pml script for this starting from scratch or from the templates general.pml or general_comments.pml (html version here).
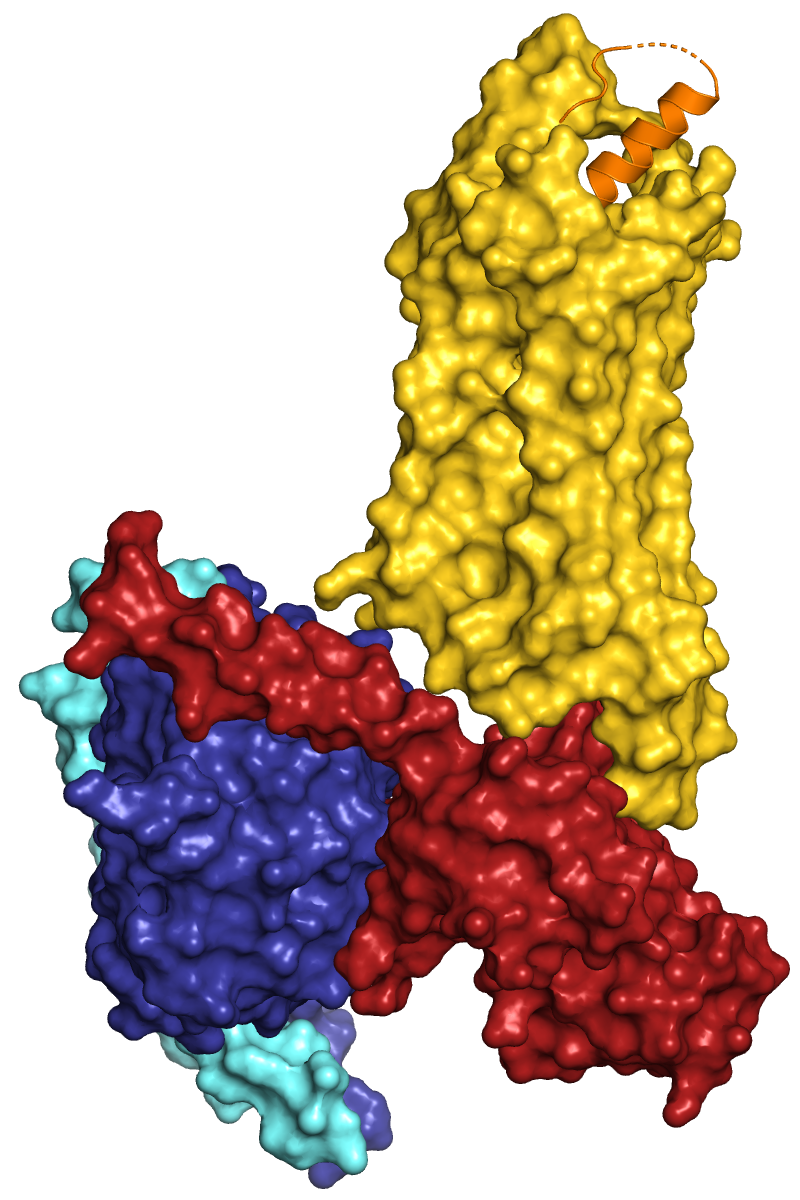
The colors I have used in the following images are: orange, gold, firebrick, deepblue, aquamarine
 |
 |
 |
| Cartoon representation of the active Y2 receptor (yellow) in
complex with a heterotrimeric G protein (Gα1 red, Gβ1
blue, Gγ2 bright blue) and the peptide agonist YY. Y2R_active_complex.pml |
Surface representation of the complex. Y2R_active_complex_surface.pml |
Y2R is shown with 50 % transparency: set transparency, 0.5, Y2 |
When you inform about the coloring of subunits, domains etc. in a figure legend, always use commonly known words that describe the used colors. Do not use the fantasy or specialised names of PyMOL or others (gold, slate, salmon, ruby, ...), even if some (like salmon) might be known to many people.
 |
 |
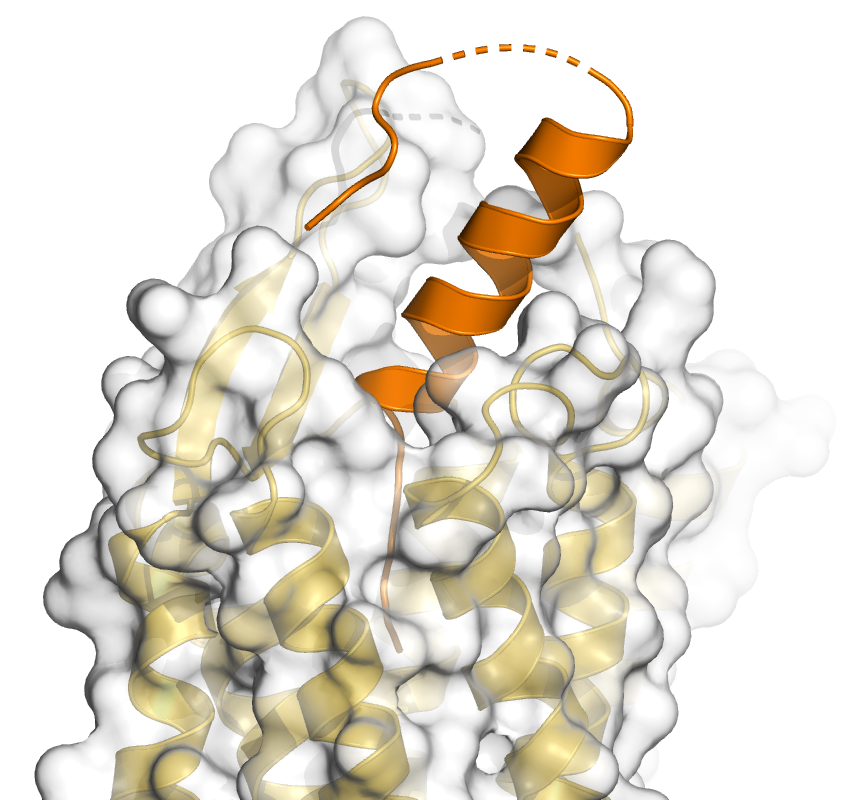 |
| The color of the Y2R cartoon has been changed to grey30. This image and the previous one have been generated with the draw command and the command "set transparency, 0.01, Galpha". This has the interesting result that the helix of the Gα1 is visible behind the Y2 surface, although it should be hidden by the red Gα1 surface. This is different when you use the ray command. Try it! Transparency and in particular the combination (superposition) of transparent objects sometimes results in surprising or unpredictable effects and you should try draw and ray, but also the different transparency modes to find a suitable solution. | Zoom on the peptide binding pocket. Ray with shadows: set ray_shadows, on Y2_active_complex_zoom_peptide.pml |
set ray_transparency_oblique, 1 |
It is quite difficult to generate a figure of the interactions of the YY peptide with the Y2 receptor due to the length of the peptide, the many interactions and in particular its interactions in the cavity of the orthosteric binding pocket. As there are a number of interactions with main chain atoms, it is probably not suitable to show only the cartoon and side chains. Therefore all residues should be displayed as sticks. Show all polar interactions between peptide and receptor via the dist command (mode=2). Next, try to find a suitable view and slap thickness to display the interactions in the orthosteric binding pocket. Omit residues in front, that are obstructing the view. If necessary, even omit some residues that are interacting with the peptide.
Back to PyMOL tutorial main page.