Peptide bond cleavage by PepA
and leucine aminopeptidase
Biological background
E. coli aminopeptidase A (PepA) und bovine lens leucine aminopeptidase
(LAP) share ~30% sequence identity. As the name implies, these exopeptidases
cleave the N-terminal amino acids from peptides. Human LAP has been shown
to catalyze postproteasomal trimming of the N-terminus of antigenic peptides
for presentation on MHC class I molecules. Here, interferon-gamma not only
promotes proteasomal cleavage but also indices LAP for N-terminal processing
of the peptides.
In addition to the aminopeptidase activity, PepA (but not LAP) has
independent
DNA-binding functions.
Hexamer structure
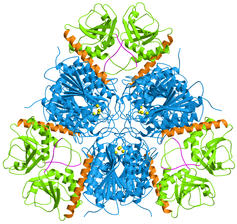 |
E. coli aminopeptidase A (PepA) is a hexameric protein
of symmetry 32, i.e. two-fold molecular axes are perpendicular to a three-fold
molecular axis. In the figure on the left, the view is along the three-fold
axis.
The C-terminal domains are shown in blue and the N-terminal domains
in green. A long helix (orange) connects the two domains. The catalytic
zinc ions are shown in yellow.
|
Compartimentalization
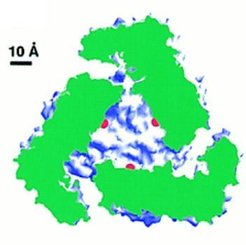 |
The aminopeptidase active sites (marked by the catalytic zinc
ions shown in red) are located in the center of the hexamer, where a large
cavity of 30 Angstrom diameter and 10 Angstrom height is formed. Access
to this cavity is provided by channels. Such a compartimentalization of
the reaction room also occurs in other proteases and in the proteasome
(see review by Larsen and Finley, 1997). In PepA and LAP the compartimentalization
ensures that the enzyme acts only on small peptides (~6 residues) and not
on proteins.
In the left figure a LAP hexamer has been cut open perpendicular to
the threefold axis. The cut protein regions are coloured green and the
rest of the protein surface in blue and white. The active sites are marked
in red.
|
The aminopeptidase active site
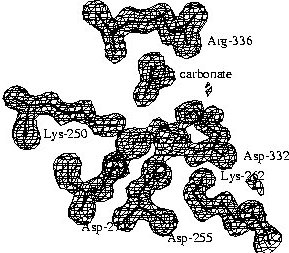 |
The electron density map of LAP at 1.6 Angstrom resolution reveals
many details of the active site. A surprising finding was the binding of
a carbonate ion next to an arginine residue. The structure of PepA, which
was determined later, and kinetic studies on wild-type PepA and mutant
variants showed that this carbonate ion is not an artifact of the crystallization
conditions, but is part of the active site and it has a functional role.
PepA is activated ~8-fold by physiological concentrations of bicarbonate
ions, i.e. that are present in the cell from dissolved carbon dioxide. |
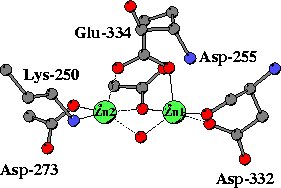 |
In the unliganded structure both zinc ions are five-coordinated,
mainly by oxygen atoms from carboxylate side chains, a peptide carbonyl
group and a water molecule. A somewhat unusual metal ligand is the lysine
residues coordinated to Zn2. |
Inhibitor binding
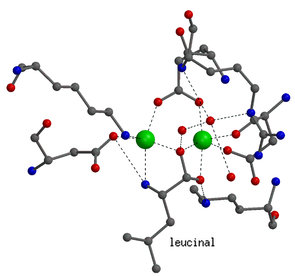 |
Details on the enzyme mechanism were obtained from the binding
of transition-state analogues to the catalytic center. Shown on the left
is the binding mode of leucinal, in which the carboxylate group of leucine
is replaced by an aldehyde group. The aldehyde group is hydrated to a gem-diol,
which mimicks the gem-diolate group of the transition-state of the reaction.
Both hydroxyl groups of the gem-diol are coordinated to the zinc ions.
In addition, the amino group of the inhibitor is also bound to one of the
zinc ions.
In the transition-state both zinc ions are six-coordinated. |
Catalytic mechanism
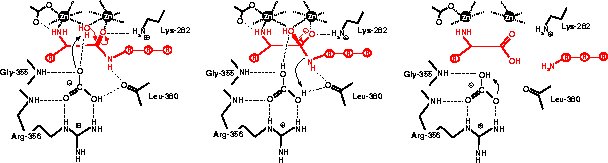
References
Larsen, C. N. & Finley, D. (1997). Protein translocation channels
in the proteasome and other proteases. Cell 91, 431-434.
Sträter, N., Sun, L., Kantrowitz, E. N. & Lipscomb, W.
N. (1999). A carbonate ion as a general base in the mechanism of peptide
hydrolysis by dizinc leucine aminopeptidase. Proc. Natl. Acad. Sci.
USA 96, 11151-11155.
Sträter, N. & Lipscomb, W. N. (1995). Two-metal ion mechanism
of bovine lens leucine aminopeptidase: active site solvent structure and
binding mode of L-leucinal, a gem-diolate transition state analogue, by
X-ray crystallography. Biochemistry 34, 14792-14800.
Sträter, N. & Lipscomb, W. N. (1995). Transition state analogue
L-leucinephosphonic acid bound to bovine lens leucine aminopeptidase: X-ray
structure at 1.65 Å resolution in a new crystal form. Biochemistry34,
9200-9210.
|



