Core Facility Mass Spectrometry
The Core Facility Mass Spectrometry offers users the infrastructure to perform mass spectrometric investigations on biomolecules, drawing on many years of experience in the field of peptide and protein analysis.
The use is open to members of the University of Leipzig, non-commercial external research institutions and, to a limited extent, also to companies. The use of the equipment can take place in self-user mode or in service mode. Further details are regulated in the usage regulations.
The costs for using the equipment are listed in the price list, which is also part of the usage regulations.
Available Equipment

ThermoFisher
Orbitrap Elite
| Ionisation | ESI, nanoESI |
| Mass analyzer | Ion Trap, Orbitrap |
| Mass range | ~50-2000 m/z |
| MS/MS | yes: MSn (n=2-10), CID and HCD |

Bruker Daltonics Microflex
| Ionisation | MALDI |
| Mass analyzer | Flugzeit linear, Flugzeit Reflektor |
| Mass range | ~500-5000 m/z reflector mode |
| ~4000-70000 m/z linear mode | |
| Target size | 48 Spots |
| MS/MS | no |

ThermoFisher
Q Exactive Plus
| Ionisation | ESI |
| Mass analyzer | Quadrupol, Orbitrap |
| Mass range | ~50-6000 m/z |
| MS/MS | yes: MS2, CID and HCD |
Intended for small molecule MS. Please Contact Dr. Matthias Gilbert or Raimund Nagel directly. Further information can be found here.
Services

Sample Preparation
Includes ZipTipping, Desalting, Conditioning and Concentrating of your Sample.
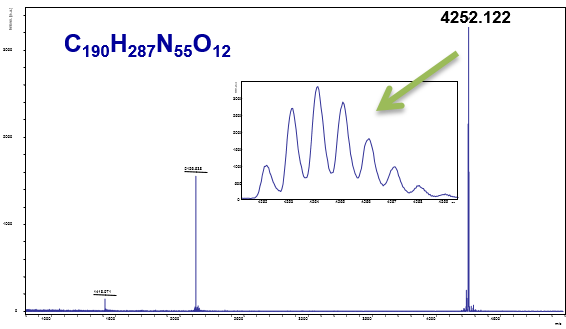
MS and MS/MS Analysis of Biomolecules
MS and MS/MS analysis of peptides and proteins, mas spectrometric characterization of nucleic acids
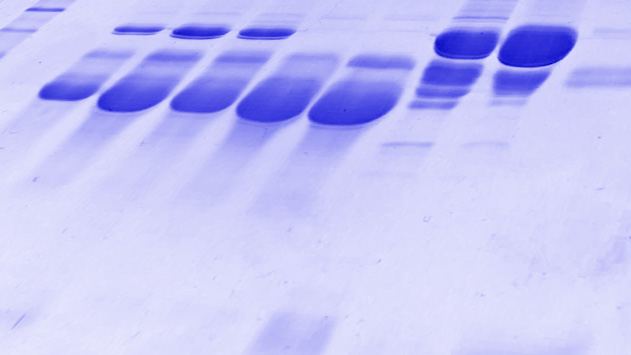
Protein identification from gel spots or solution
Includes the identification of proteins after enzymatic digestion of gel bands, the identification of post-translational modifications and the characterization of disulfide bond formation.
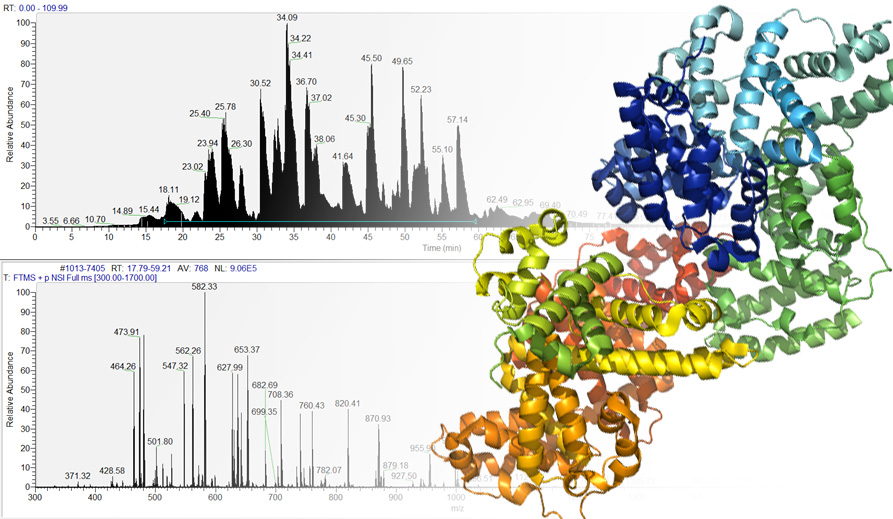
Analyzing complex data
We offer help on analyzing complex data obtained from proteolytic digestion, protein-protein interaction studies and crosslinking.
Contact us for further information...
