A7 – Molecular modeling and simulation of glycosaminoglycans (GAGs)-protein interactions
This project focuses on developing and applying computational approaches to study GAG-protein interactions in order to better understand their requirements for affinity and specificity and to characterize the molecular mechanisms responsible for their function. We use the knowledge acquired to stablish structure-function relationships theoretical models and to propose rationale for customized specificity in order to guide the engineering of new biomaterials and thus be able to tune the interplay GAG/target-protein/receptor for functional purposes.
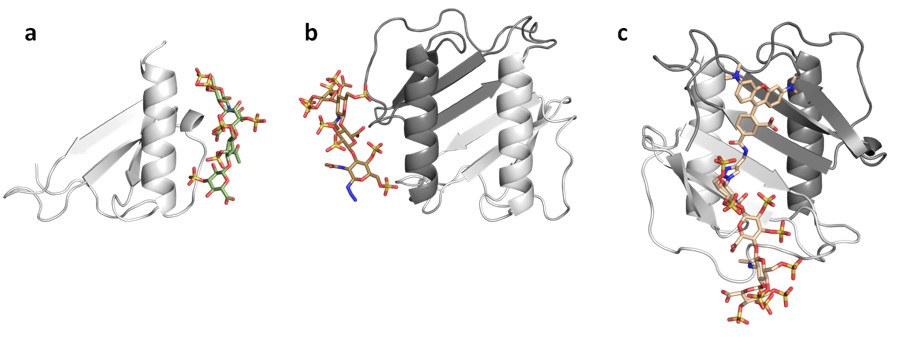
A great part of our work consists on applying computational tools in cooperation with experimentalists to investigate the interaction of a variety of GAGs with extracellular matrix target protein systems. We establish theoretical models for the analysis of the conformational properties of GAG derivatives, the location of their binding sites, the characterization of relevant target protein residues, as well as the interplay with their receptors. This integrative endeavour strengths the predictive value of our theoretical models, notably assists in the establishment of working hypothesis and their assessment, and advances our knowledge on GAG-protein molecular recognition and its functional implications in the investigated systems. Noteworthy results of our work are, for instance, the obtained new structural and functional insights into GAG-sclerostin interactions in bone (together with project B2) and into the molecular mechanism used by sulfated GAGs to exploit BMP-2 conformational plasticity (Fig. 1) and alter its receptor interactions (together with project A3).
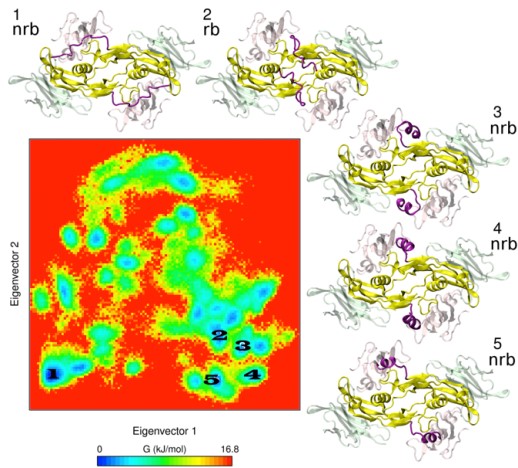
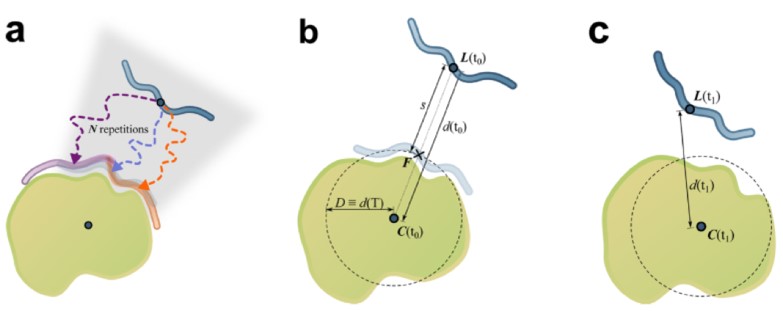
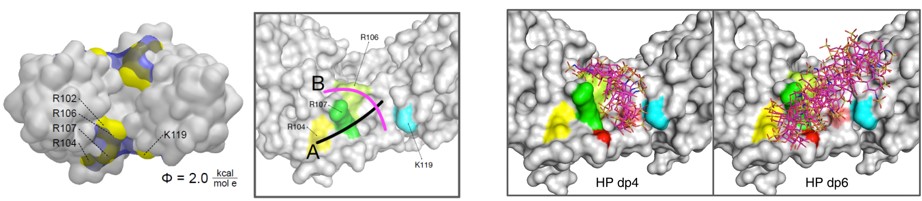
We also develop computational methodology for the prediction and study of GAG-protein interactions. For instance, we have established a new molecular dynamics (MD)-based approach for molecular docking (DMD; Fig. 2), which takes GAG flexibility and explicit solvent into account and is optimized for GPU technology for intensive calculations. This method has allowed the characterization of GAG derivatives binding to a diverse collection of extracellular matrix proteins (Fig. 3-4).
Our studies will help in the design of building blocks for chemical and modular synthesis of GAGs-derivatives and matrix scaffolds in biomaterials to promote bio-specific cell behaviour.
Publications
- Balamurugan K, Koehler L, Dürig JN, Hempel U, Rademann J, Hintze V, Pisabarro MT. Structural insights into the modulation of PDGF/PDGFR-β complexation by hyaluronan derivatives. Biological Chemistry, vol. 402, no. 11, 2021, pp. 1441-1452. https://doi.org/10.1515/hsz-2021-0173
- Ruiz-Gómez G, Vogel S, Möller S, Pisabarro MT, Hempel U. Glycosaminoglycans influence enzyme activity of MMP2 and MMP2/TIMP3 complex formation – Insights at cellular and molecular level. Sci Rep 2019;9:4905,1-15. doi:10.1038/s41598-019-41355-2
- Koehler L, Ruiz-Gómez G, Balamurugan K, Rother S, Freyse J, Möller S, Schnabelrauch M, Köhling S, Djordjevic S, Scharnweber D, Rademann J, Pisabarro MT, Hintze V. Dual Action of Sulfated Hyaluronan on Angiogenic Processes in Relation to Vascular Endothelial Growth Factor-A. Sci Rep. 2019 Dec 2;9(1):18143. doi: 10.1038/s41598-019-54211-0.
- Nordsieck K, Baumann L, Hintze V, Pisabarro MT, Schnabelrauch M, Beck-Sickinger AG, Samsonov SA. The effect of interleukin-8 truncations on its interactions with glycosaminoglycans. Biopolymers. 2018; 109:e23103.
- Wodtke R, Hauser C, Ruiz-Gómez G, Jäckel E, Bauer D, Lohse M, Wong A, Pufe J, Ludwig FA, Fischer S, Hauser S, Greif D, Pisabarro MT, Pietzsch J, Pietsch M, Löser R. N?-Acryloyllysine Piperazides as Irreversible Inhibitors of Transglutaminase 2: Synthesis, Structure-Activity Relationships, and Pharmacokinetic Profiling. J Med Chem. 2018, 61:4528-60.
- Köhler L, Samsonov S, Rother S, Vogel S; Köhling S, Möller S, Schnabelrauch M, Rademann J, Hempel U, Pisabarro MT, Scharnweber D, Hintze V. Sulfated hyaluronan derivatives modulate TGF-ß1:receptor complex formation: Possible consequences for TGF- ß1 signaling. Scientific Reports. 2017;7:1210.
- Jouy F, Lohmann N, Wandel E, Ruiz-Gómez G, Pisabarro MT, Beck-Sickinger AG, Schnabelrauch M, Moeller S, Simon JC, Kalkhof S, von Bergen M, Franz S. Sulfated hyaluronan attenuates inflammatory signaling pathways in 1 macrophages involving induction of antioxidants. Proteomics. 2017. [Epub ahead of print]
- Rother S, Samsonov S, Möller S, Schnabelrauch M, Rademann J, Blaszkiewicz J, Köhling S, Waltenberger J, Pisabarro MT, Scharnweber D, Hintze V. Sulfate hyaluronan alters endothelial cell activation in vitro by controlling the biological activity of the angiogenic factors vascular endothelial growth factor-A and tissue inhibitor of metalloproteinase-3. ACS Appl Mater Interfaces. 2017; 9:9539-9550.
- Rother S, Samsonov SA, Hofmann T, Blaszkiewicz J. Köhling S. Moeller S, Schnabelrauch M, Rademann J, Kalkhof S, von Bergen M, Pisabarro MT, Scharnweber D, Hintze V. Structural and functional insights into the interaction of sulfated GAGs with tissue inhibitor of metallopro-teinase-3 – A possible regulatory role on extracellular matrix homeostasis. Acta Biomater. 2016;45:143-154.
- Ruiz-Gómez G. Hawkins JC, Philipp J, Künze G, Wodtke R, Löser R, Fahmy K, Pisabarro MT. Rational structure-based rescaffolding approach to de novo design of interleukin 10 (IL-10) receptor-1 mimetics. Plos One. 2016;11:e0154046.
- Babik S, Samsonov SA, Pisabarro MT. Computational drill down on FGF1-heparin interactions through methodological evaluation. Glycoconjugate Journal. 2016;34:427-440.
- Samsonov SA, Pisabarro MT. Computational analysis of interactions in structurally available protein-glycosaminoglycan complexes. Glycobiology. 2016;26:850-61.
- Rother S, Samsonov SA, Hempel U, Vogel S, Moeller S, Blaszkiewicz J, Köhling S, Schnabelrauch M, Rademann J, Pisabarro MT, Hintze V, Scharnweber D. Sulfated hyaluronan alters the interaction profile of TIMP-3 with the endocytic receptor LRP-1 cluster II and IV and increases the extracellular TIMP-3 level of human bone marrow stromal cells. Biomacromolecules. 2016; 17:3252-3261.
- Rother S, Samsonov SA, Hofmann T, Blaszkiewicz J, Köhling S, Moeller S, Schnabelrauch M, Rademann J, Kalkhof S, von Bergen M, Pisabarro MT, Scharnweber D, Hintze V. Structural and functional insights into the interaction of sulfated glycosaminoglycans with tissue inhibitor of metalloproteinase-3 – a possible regulatory role on extracellular matrix homeostasis. Acta Biomater. 2016, 45:143-54.
- Picke AK, Salbach-Hirsch J, Hintze V, Rother S, Rauner M, Kascholke C, Möller S, Bernhardt R, Rammelt S, Pisabarro MT, Ruiz-Gómez G, Schnabelrauch M, Schulz-Siegmund M, Hacker MC, Scharnweber D, Hofbauer C, Hofbauer LC. Sulfated hyaluronan improves bone regeneration of diabetic rats by binding sclerostin and enhancing osteoblast function. Biomaterials. 2016;96:11-23.
- Panitz N, Theisgen S, Samsonov SA, Gehrke JP, Baumann L, Bellmann-Sickert K, Köhling S, Pisabarro MT, Rademann J, Huster D, Beck-Sickinger AG. The structural investigation of glycosaminoglycan binding to CXCL12 displays distinct interaction sites. Glycobiology. 2016;26:1-13.
- Gehrcke JP, Pisabarro MT. Identification and characterization of a GAG binding site on Inter-leukin-10 via molecular simulation methods. J Mol Graph Model. 2015;62:97-104.
- Salbach-Hirsch J, Samsonov SA, Hintze V, Hofbauer C, Picke AK, Rauner M, Gehrcke JP, Moeller S, Schnabelrauch M, Scharnweber D, Pisabarro MT, Hofbauer LC. Structural and functional insights into sclerostin-GAG interactions in bone. Biomaterials. 2015; 67:335-45.
- Anselmi M, Pisabarro MT. Exploring multiple binding modes using confined replica exchange molecular dynamics. Journal of Chemical Theory and Computation. 2015;11:3906-3918.
- Hofmann T, Samsonov SA, Pichert A, Lemmnitzer K, Schiller J, Huster D, Pisabarro MT, von Bergen M, Kalkhof S. Structural analysis of the interleukin-8/glycosaminoglycan interactions by amide hydrogen/deuterium exchange mass spectrometry. Methods. 2015. [Epub ahead of print]
- Samsonov SA, Bichmann L, Pisabarro MT. Coarse-grained model of glycosaminoglycans. J Chem Inf Model, 2015; 55(1):114-24.
Atkovska K, Samsonov SA, Paszkowski-Rogacz M, Pisabarro MT. Multipose binding in mo-lecular docking. Int J Mol Sci. 2014;15:2622-45. - Hintze V, Samsonov SA, Anselmi M, Möller S, Becher J, Schnabelrauch M, Scharnweber D, Pisabarro MT. Sulfated glycosaminoglycans exploit the conformational plasticity of bone morphogenetic protein-2 (BMP-2) and alter the interaction profile with its receptor. Biomacromolecules, 2014; 15(8):3083-92.
- Scharnweber D, Hübner L, Rother S, Hempel U, Anderegg U, Samsonov SA, Pisabarro MT, Hofbauer L, Schnabelrauch M, Franz S, Simon JC, Hintze V. Glycosaminoglycan derivates: promising candidates for the design of functional biomaterials. J Mater Sci: Mater Med;2015;26:232.
- Samsonov SA, Theisgen S, Riemer T, Huster D, Pisabarro MT. Glycosaminoglycan monosac-charide blocks analysis by quantum mechanics, molecular dynamics and nuclear magnetic res-onance. BioMed Res Int. 2014;2014:808071.
- Kuenze G, Gehrcke JP, Pisabarro MT, Huster D. NMR characterization of the binding properties and conformation of glycosaminoglycans interacting with interleukin-10. Glycobiology, 2014;24:1036-49.
- Samsonov S, Gehrcke JP, Pisabarro MT. Flexibility and explicit solvent in molecular dynamics-based docking of protein-glycosaminoglycan systems. J Chem Inf Model, 2014;54:582-92.
- Atkovska K, Samsonov SA, Paszkowski-Rogacz M, Pisabarro MT. Multipose binding in molecular docking. International Journal of Molecular Sciences. 2014;15:2622-2645.
- Samsonov SA, Pisabarro MT. Importance of IdoA and IdoA(2S) ring conformations in computational studies of GAG-protein interactions. Carbohydr Res. 2013;381:133-137.
- Salbach-Hirsch J, Kraemer J, Rauner M, Samsonsov SA, Pisabarro MT, Moeller S, Schnabelrauch M, Scharnweber D, Hofbauer LC, Hintze V. The promotion of osteoclastogenesis by sulfated hyaluronan through interference with osteoprotegerin and receptor activator of NF-?-B ligand/osteoprotegerin complex formation. Biomaterials 2013; 34(31):7653-61.
- van der Smissen A, Samsonsov SA, Hintze V, Scharnweber D, Moeller S, Schnabelrauch M, Pisabarro MT, Anderegg U. Artificial extracellular matrix composed of collagen l and high-sulfated hyaluronan interferes with TGF?1 signaling and prevents TGF?1 induced myofribroblast differentiation. Acta Biomaterials 2013;2:226-36.
- Möbius K, Nordsieck K, Pichert A, Samsonsov SA, Thomas L, Schiller J, Kalkhof S, Pisabarro MT, Beck-Sickinger AG, Huster D. Investigation of lysine side chain interactions of interleukin-8 with heparin and other glycosaminoglycans studied by a methylation-NMR approach. Glycobiology. 2013;23:1260-9.
- Sage J, Mallevre F, Barbarin-Costes F, Samsonov SA, Gehrcke JP, Pisabarro MT, Perrier E, Schnebert S, Roget A, Livache T, Nizard C, Lalmanach G, Lecaille F. Binding of chondroitin 4-sulfate to cathepsin S regulates its enzymatic activity. Biochemistry. 2013;52:6487-98.
- Nordsieck K, Pichert A, Samsonov SA, Thomas L, Berger C, Pisabarro MT, Huster D, Beck-Sickinger AG. Residue 75 of Interleukin-8 is Curicial for the Interaction with Glycosaminoglycans. ChemBioChem. 2012;13:2558-66.
- Pichert A, Samsonov SA, Theisgen S, Thomas L, Baumann L, Schiller J, Beck-Sickinger AG, Huster D, Pisabarro MT. Characterization of the interaction of the interaction of Interleukin-8 with hyaluronan, chondroitin sulfate, dermatan sulfate, and their sulfated derivatives by spectroscopy and molecular modelling. Glycobiology. 2012;22:134-45.
- Teyra J, Samsonov SA, Schreiber S, Pisabarro MT. SCOWLP update: 3D classification of protein-protein, -peptide, -saccaride, and -nucleid acid interactions, and structure-based binding inferences across folds. BMC Bioinformatics. 2011:12:398.
- Samsonov SA, Teyra J, Pisabarro MT. Docking glycosaminoglycans to proteins: analysis of solvent inclusion. J Comput Aided Mol Des. 2011;25:477-89.
Contact
Dr. María Teresa Pisabarro
Group Leader Structural Bioinformatics and Bioinformatics
Dresden University of Technology
Biophysics/BIOTEC
Tatzberg 47 – 51, 01307 Dresden
Phone: +49 (0)351 463-40070
E-Mail: mayte.pisabarro (at) biotec.tu-dresden.de
Web: biotec.tu-dresden.de/research/pisabarro.html
