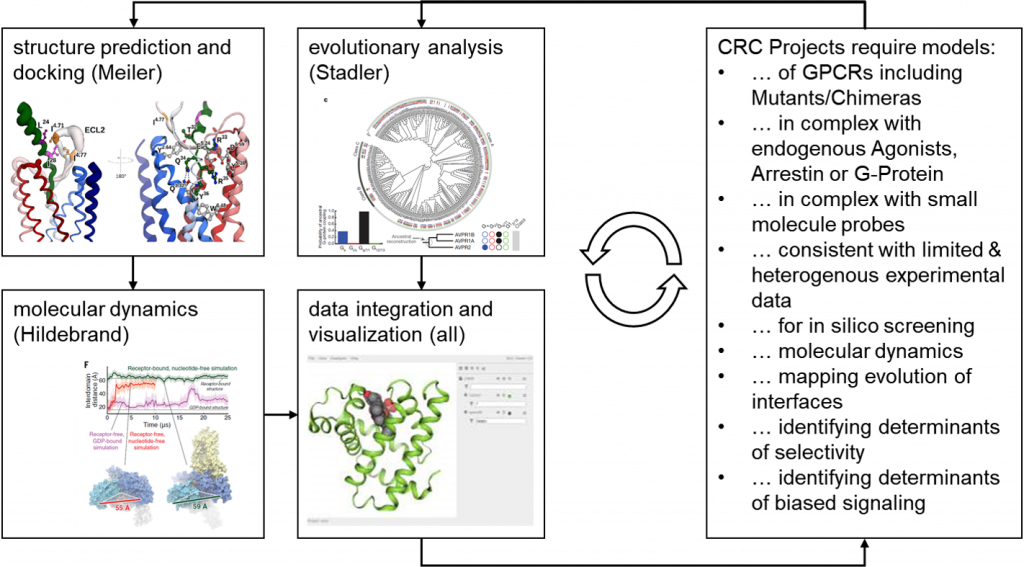
Project Z04-INF – Data management and computational models of structure and dynamics, and evolution of GPCRs
Z04-INF will support the CRC by providing a comprehensive computational analysis of the receptors and their interacting partners, and will also cover the necessary aspects of data management. In an iterative loop, we will build computational models consistent with experimental data, combining Rosetta modeling, molecular dynamics simulation, allosteric network analysis, and evolutionary analysis. From these models, we will derive testable hypothesis for the next iteration of experimental validation in the respective projects. In addition, a dedicated INF component will be added, focusing on metadata collection and the creation of a non-invasive platform for data exchange.
Contact

Prof. Dr. Peter Hildebrand (Project Leader)
Leipzig University, Faculty of Medicine
Institute of Medical Physics and Biophysics
Härtelstraße 16 – 18, D-04107 Leipzig
Phone +49 341 97 15705
E-Mail
Web uni-leipzig.de/prof-dr-peter-w-hildebrand

Prof. Dr. Jens Meiler (Project Leader)
Leipzig University, Faculty of Medicine
Institute for Drug Development
Liebigstraße 19, D-04103 Leipzig
Phone +49 341 97 11801
E-Mail
Web https://meilerlab.org/
Web https://www.uniklinikum-leipzig.de/einrichtungen/wirkstoffentwicklung

Dr. Franziska Reinhardt (PostDoc)
Leipzig University, Faculty of Mathematics and Computer Science
Institute of Computer Science
Härtelstraße 16 – 18, D-04107 Leipzig

Dr. Hossein Batebi (PostDoc)
Leipzig University, Faculty of Mathematics and Computer Science
Institute of Computer Science
Härtelstraße 16 – 18, D-04107 Leipzig
Resources
We focus on:
- Structural intermediates, complexes
- Combining MD simulations with evolutionary analysis
- Performing MD simulations of the membrane-embedded proteins
- Usage of paramagnetic restranst in structre determination
Techniques
- computational models
- MD Simulations: Classical Molecular Dynamics, QM/MM, QMD, DFT, Enhanced Sampling, AMBER, Gromacs, VMD
- evolutionary analysis
Publications
Conflitti P, Lyman E, Sansom MSP, Hildebrand PW, Gutiérrez-de-Terán H, Carloni P, Ansell TB, Yuan S, Barth P, Robinson AS, Tate CG, Gloriam D, Grzesiek S, Eddy MT, Prosser S, Limongelli V. Functional dynamics of G protein-coupled receptors reveal new routes for drug discovery. Nat Rev Drug Discov. 2025 Jan 2. doi: 10.1038/s41573-024-01083-3. Epub ahead of print. PMID: 39747671.
Ertelt M, Moretti R, Meiler J, Schoeder CT. Self-supervised machine learning methods for protein design improve sampling but not the identification of high-fitness variants. Sci Adv. 2025 Feb 14;11(7):eadr7338. doi: 10.1126/sciadv.adr7338. Epub 2025 Feb 12. PMID: 39937901; PMCID: PMC11817935.
Lin X, Su Z, Liu YL, Liu J, Kuang X, Cummings PT, Spencer-Smith J, Meiler J. SuperMetal: A Generative AI Framework for Rapid and Precise Metal Ion Location Prediction in Proteins. bioRxiv [Preprint]. 2025 Mar 25:2025.03.21.644685. doi: 10.1101/2025.03.21.644685. PMID: 40196543; PMCID: PMC11974720.
Liu Y, Moretti R, Wang Y, Dong H, Yan B, Bodenheimer B, Derr T, Meiler J. Advancements in Ligand-Based Virtual Screening through the Synergistic Integration of Graph Neural Networks and Expert-Crafted Descriptors. J Chem Inf Model. 2025 May 14. doi: 10.1021/acs.jcim.5c00822 . Epub ahead of print. PMID: 40365985 .
Marlow B, Vogel A, Kuenze G, Pankonin M, Reinhardt F, Stadler PF, Hildebrand PW, Meiler J. Cholesterol Allosteric Modulation of the Oxytocin Receptor. Biophys J. 2025 Apr 29:S0006-3495(25)00250-4. doi: 10.1016/j.bpj.2025.04.023. Epub ahead of print. PMID: 40308029.
Pérez-Hernández G, Hildebrand PW. mdciao: Accessible Analysis and Visualization of Molecular Dynamics Simulation Data. PLoS Comput Biol. 2025 Apr 21;21(4):e1012837. doi: 10.1371/journal.pcbi.1012837 . PMID: 40258031 ; PMCID: PMC12011235.
Schermeng T, Fürll A, Liessmann F, von Bredow L, Stichel J, Weaver CD, Tretbar M, Meiler J, Beck-Sickinger AG. Similar Binding Mode of a 5-Sulfonylthiouracil Derivative Antagonist at Chemerin Receptors CMKLR1 and GPR1. J Med Chem. 2025 May 16. doi: 10.1021/acs.jmedchem.5c00135. Epub ahead of print. PMID: 40377914.
Scott T, Smethurst CAP, Westermaier Y, Mayer M, Greb P, Kousek R, Biberger T, Bader G, Jandova Z, Schmalhorst PS, Fuchs JE, Magarkar A, Hoenke C, Gerstberger T, Combs SA, Pape R, Phul S, Kothiwale S, Bergner A; Foldit Players; Waterson AG, Weinstabl H, McConnell DB, Meiler J, Böttcher J, Moretti R. Drugit: crowd-sourcing molecular design of non-peptidic VHL binders. Nat Commun. 2025 Apr 14;16(1):3548. doi: 10.1038/s41467-025-58406-0. PMID: 40229246; PMCID: PMC11997059.
Seufert F, Pérez-Hernández G, Pándy-Szekeres G, Guixà-González R, Langenhan T, Gloriam DE, Hildebrand PW. Generic residue numbering of the GAIN domain of adhesion GPCRs. Nat Commun. 2025 Jan 2;16(1):246. doi: 10.1038/s41467-024-55466-6. PMID: 39747076; PMCID: PMC11697300.
Zlobin A, Maslova V, Beliaeva J, Meiler J, Golovin A. Long-Range Electrostatics in Serine Proteases: Machine Learning-Driven Reaction Sampling Yields Insights for Enzyme Design. J Chem Inf Model. 2025 Feb 24;65(4):2003-2013. doi: 10.1021/acs.jcim.4c01827. Epub 2025 Feb 10. PMID: 39928564; PMCID: PMC11863386.
Batebi H, Pérez-Hernández G, Rahman SN, Lan B, Kamprad K, Shi M, Speck D, Tiemann JKS, Guixà-González R, Reinhardt F, Stadler PF, Papasergi-Scott MM, Skiniotis G, Scheerer P, Kobilka BK, Mathiesen JM, Liu X, Hildebrand PW. Mechanistic insights into G-protein coupling with an agonist-bound G-protein-coupled receptor. Nature Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01334-2 .
Eisenhuth P, Liessmann F, Moretti R, Meiler J. REvoLd: Ultra-Large Library Screening with an Evolutionary Algorithm in Rosetta. ArXiv [Preprint]. 2024 Apr 26:arXiv:2404.17329v1.
Klemm P, Stadler PF, Lechner M. Proteinortho6: pseudo-reciprocal best alignment heuristic for graph-based detection of (co-)orthologs. Front Bioinform. 2023 Dec 13;3:1322477.
Sala D, Batebi H, Ledwitch K, Hildebrand PW, Meiler J. Targeting in silico GPCR conformations with ultra-large library screening for hit discovery. Trends Pharmacol Sci. 2023; 44:150-61.
Sala D, Hildebrand PW, Meiler J. Biasing AlphaFold2 to predict GPCRs and Kinases with user-defined functional or structural properties. Front Mol Biosci. 2023; 10:1121962.
Schmidt P, Vogel A, Schwarze B, Seufert F, Licha K, Wycisk V, Kilian W, Hildebrand PW, Mitschang L. Towards Probing Conformational States of Y2 Re-ceptor Using Hyperpolarized 129Xe NMR. Molecules. 2023; 28:1424
Staritzbichler R, Ristic N, Stapke T, Hildebrand P. SmoothT – a server constructing low energy pathways from conformational ensembles for interactive visualization and enhanced sampling. Bioinformatics. 2023 Apr 5:btad176.
Schaller D, Lafond M, Stadler PF, Wieseke N, Hellmuth M. Indirect identification of horizontal gene transfer. Journal of Mathematical
Biology. 2021; 83:10. https://doi.org/10.1007/s00285-021-01631-0.
Kasmanas JC. Bartholomäus A, Corrêa FB, Tal T, Jehmlich N, Herberth Gunda, von Bergen M, Stadler PF, de Carvalho ACPLF, da Rocha UN. HumanMetagenomeDB: a public repository of curated and standardized metadata for human metagenomes. Nucleic Acids Res. 2021; 49:D743-D750.
Koehler Leman J, Lyskov S, Lewis SM, Adolf-Bryfogle J, Alford RF, Barlow K, Ben-Aharon Z, Farrell D, Fell J, Hansen WA, Harmalkar A, Jeliazkov J, Kuenze G, Krys JD, Ljubetič A, Loshbaugh AL, Maguire J, Moretti R, Mulligan VK, Nance ML, Nguyen PT, Ó Conchúir S, Roy Burman SS, Samanta R, Smith ST, Teets F, Tiemann JKS, Watkins A, Woods H, Yachnin BJ, Bahl CD, Bailey-Kellogg C, Baker D, Das R, DiMaio F, Khare SD, Kortemme T, Labonte JW, Lindorff-Larsen K, Meiler J, Schief W, Schueler-Furman O, Siegel JB, Stein A, Yarov-Yarovoy V, Kuhlman B, Leaver-Fay A, Gront D, Gray JJ, Bonneau R. Ensuring scientific reproducibility in bio-macromolecular modeling via extensive, automated benchmarks. Nat Commun. 2021; 12:6947.
Nunn A, Can SN, Otto C, Fasold M, Diez Rodrigues B, Fernandez-Pozo N,Rensing SA, Stadler PF, Langenberger D. EpiDiverse Toolkit: a pipeline suite for the analysis of bisulfite sequencing data in ecological plant epigenetics. Nucleic Acids Res. Genom Bioinf. 2021; 3:lqab106.
Rudolf S, Kaempf K, Vu O, Meiler J, Beck-Sickinger AG, Coin I. Binding of Natural Peptide Ligands to the Neuropeptide Y5 Receptor. Angew Chem Int Ed Engl. 2021 Nov 25. doi: 10.1002/anie.202108738 . PMID:34822209.
Staritzbichler R, Ristic N, Goede A, Preissner R, Hildebrand PW. Voronoia 4-ever. Nucleic Acids Res. 2021 Jun 9:gkab466. doi: 10.1093/nar/gkab466. Epub ahead of print. PMID: 34107038.
Koehler Leman J, Weitzner BD, Renfrew PD, Lewis SM, Moretti R, Watkins AM, Mulligan VK, Lyskov S, Adolf-Bryfogle J, Labonte JW, Krys J; RosettaCommons Consortium, Bystroff C, Schief W, Gront D, Schueler-Furman O, Baker D, Bradley P, Dunbrack R, Kortemme T, Leaver-Fay A, Strauss CEM, Meiler J, Kuhlman B, Gray JJ, Bonneau R. Better together: Elements of successful scientific software development in a distributed collaborative community. PLoS computational biology. 2020; 16:e1007507.
Lem Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, Alam N, Alford RF, Aprahamian M, Baker D, Barlow KA, Barth P, Basanta B, Bender BJ, Blacklock K, Bonet J, Boyken SE, Bradley P, Bystroff C, Conway P, Cooper S, Correia BE, Coventry B, Das R, De Jong RM, DiMaio F, Dsilva L, Dunbrack R, Ford AS, Frenz B, Fu DY, Geniesse C, Goldschmidt L, Gowthaman R, Gray JJ, Gront D, Guffy S, Horowitz S, Huang PS, Huber T, Jacobs TM, Jeliazkov JR, Johnson DK, Kappel K, Karanicolas J, Khakzad H, Khar KR, Khare SD, Khatib F, Khramushin A, King IC, Kleffner R, Koepnick B, Kortemme T, Kuenze G, Kuhlman B, Kuroda D, Labonte JW, Lai JK, Lapidoth G, Leaver-Fay A, Lindert S, Linsky T, London N, Lubin JH, Lyskov S, Maguire J, Malmström L, Marcos E, Marcu O, Marze NA, Meiler J, Moretti R, Mulligan VK, Nerli S, Norn C, Ó’Conchúir S, Ollikainen N, Ovchinnikov S, Pacella MS, Pan X, Park H, Pavlovicz RE, Pethe M, Pierce BG, Pilla KB, Raveh B, Renfrew PD, Burman SSR, Rubenstein A, Sauer MF, Scheck A, Schief W, Schueler-Furman O, Sedan Y, Sevy AM, Sgourakis NG, Shi L, Siegel JB, Silva DA, Smith S, Song Y, Stein A, Szegedy M, Teets FD, Thyme SB, Wang RY, Watkins A, Zimmerman L, Bonneau R. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nature methods. 2020; 17:665-80.