Research Data Management of the SFB1423
Following existing practice, significant findings resulted from work conducted under the SFB1423 will be promptly prepared and submitted for peer-reviewed journal publication, with authorship that accurately reflects the contributions of those involved. Details on specific aspects regarding data management are described below.
Research Data Policy
The SFB1423 conforms to the DFG Guidelines for Research Data Management and in according to the regulations of the respective faculties according to their official duties, e.g. Leipzig University “Principles for the management of research data”.
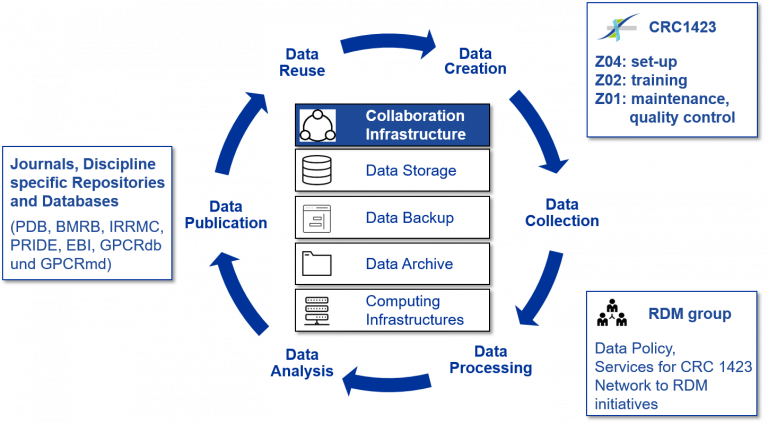
Research Data Management Plan
Data generated within the SFB1423 can include
- computer algorithms and source code including appropriate documentation,
- computational protocols to apply said algorithms to research questions,
- experimental protocols for sample production and preparation (peptide synthesis, protein expression, cell and animal studies),
- documentation of sample preparation including but not limited to vectors, gels, chromatograms, im-ages, cell culture conditions, animal studies
- data from sample analysis with biophysical methods including but not limited to crystallography, UV-VIS-, NMR-, EPR-spectroscopy, mass spectrometry, fluorescence spectroscopy, bioluminescence spectrometry and calorimetry,
- data from studies with cells, stably/transiently transfected cell lines,
- data from studies with C. elegans or D. melanogaster,
- trajectories from MD simulations and related analysis data.
Data as enumerated above will be stored as hardcopies – e.g. laboratory notebook, instrument outputs – as well as in readily usable electronic file formats, including, but not limited to website which allows its users to add and modify data (wiki, confluence), source code (cpp, h) text (pdf, ascii, office), and images (jpeg, png). Computer algorithms represented as source code are managed by a version control system (VCS) system. On this basis, functional extensions, bug fixes and other changes are documented in a transparent and traceable way so that several developers can work on the same code base at the same time. UL operates a git infrastructure for this purpose that supports the research and development process with additional project management and collaboration functionalities. Our source code will be doc-umented using an industry-standard documentation system (doxygen). Computational protocols used to generate data for publication will be formatted as a ‘protocol capture’, a directory that contains all scripts and command files, sample input data, as well as sample output data together with a detailed description ‘readme’ on how to reproduce published results.
Experimental protocols for sample synthesis, analysis and preparation will be shared and stored within the SFB1423 on the laboratories confluence pages – a wiki-like system that allows maintenance and sharing of data within the laboratory and with collaborators. Sample preparation will be primarily docu-mented in laboratory notebooks (hardcopy) with key results being also documented on the confluence pages. Data from sample analysis with biophysical methods will be collected and stored as raw instrument output on the computer file system as well as processed data on the confluence pages.
All personnel within the CRC1423 is required to maintain complete records of all data produced and protocols on how data were collected in the above detailed formats. Electronic data is stored on a scalable, high-performance network attached storage system (NAS) of the URZ (UL University Computing Centre), the MDC, MLU Halle or Charité. A redundant array of independent disks (RAID) system ensures that upon failure of individual disks no data is corrupted or deleted. In addition, data is backed-up nightly on a tape back-up system. After completion and publication of a research project data is kept for at least one year before it is transferred into a tape archiving system where it is long-term stored.
In addition to these centralized solutions for data storage, back-up and archiving personnel on the project maintain laboratory notebooks, store electronic data on personal computers, and maintain copies on common disk space. All laboratory personnel generate electronic hardcopies in form of DVDs or CDs periodically or external drives. Upon leaving the laboratory all personnel passes laboratory notebooks, DVDs, and CDs to the PI for storage.
The NMR groups use the LOGS system (https://logs-repository.com/), a research data management plat-form developed for the needs of NMR spectroscopists. The system provides a solution that secures primary raw data on cloud memory space. With LOGS, NMR data is retrieved from the NMR spectrometers auto-matically; data is tagged and required to be linked to a note in the notebook of the respective operator and can also be linked to processed data, diagrams, tables, and charts etc. that appear in publications. Data are stored for at least 10 years even if the company goes bankrupt.
MS data are archived on a regular basis at the HRZ (MLU Halle).
The papers and supporting information will all be accessible on the appropriate publisher’s website in pdf or html format, and when permissible by the publisher, will also be accessible on the central CRC website and the laboratory websites. Generally, no reasons exist to not share generated data and metadata. In instances when intellectual property may arise that might warrant a patent or other form of protection, the PI will work with the Technology Transfer office to ensure that the technology remains widely available to the research community. Generally, data will be opened to wider use without reserving any exclusive rights to the data collector making provisions only for appropriate protection of privacy, confidentiality, se-curity, intellectual property, or other rights or requirements.
Computational programs developed will be accessible to the public via server(s) hosted by the PI and/or also as executable via ROSETTACOMMONS or BCLCOMMONS. Both commons are communities of researchers at various institutions, including Leipzig University and Vanderbilt University, that develop and implement improvements to the ROSETTA and BCL software. The commons use a shared code base to ensure that methods produced in different labs are fully compatible and seamlessly integrated into the computer program. The commons make the code freely available for modification under the condition that any improvements or additions are contributed back into the common source code base. ROSETTA and BCL are licensed for free to users at academic and nonprofit institutions. Commercial entities can use ROSETTA and BCL by paying a license fee. Sequence analysis tools and related software will be made available to the community as open source via github and/or the PIs’ websites.
Selected data will be published in online databases. Specifically, atomic coordinates of models will be published in ModBase when appropriate or made available upon request. Atomic coordinates of exper-imental structures will be submitted to the Protein Data Bank (PDB). Other experimental data will be sub-mitted to the respective community repositories such as the Biological Magnetic Resonance Bank for NMR data (BMRB). Trajectory data from MD simulations are made available through MDsrv sessions that will be deposited at www.proteinformatics.de (Hildebrand laboratory), allowing interactive visualization of the tra-jectories directly within standard web browsers. GPCR related simulation data can in addition be deposited at the GPCRmd website: http://www.gpcrmd.org which uses interactive MDsrv visualization along with re-lated analysis. Collected GPCR sequence data will be deposited at the GPCRdb: https://gpcrdb.org.
There will be a close collaboration of Z01–Z04 within the SFB1423 with the research Data Man-agement Team (RDM) at Leipzig University to ensure Data Management. Z04 will set up the confluence pages (wiki) for all members of the SFB1423. There will be a continuous training of students and new members within special courses of Z02 (MGK) in close collaboration with the RDM.
Standardized protocols worked out in Z03 will be listed on the confluence page and all data, obtained for peptides and proteins in Z03 will be available to all projects. Z01 will supervise data management, regularly check and discuss with PIs to improve handling, to guarantee quality control, negotiate with URZ for storage and maintenance and ensure collaboration with RDM.