About our research
The CRC aims to elucidate structural hallmarks in various states of activation, which are linked to GPCR func-tion. A broad spectrum of methods will be applied synergistically: computational techniques, structural meth-ods, site-directed mutagenesis, crosslinking and mass spectrometry (MS), as well as functional cell-based analyses. The structural dynamics of peptide GPCRs and aGPCRs will be compared to those of the well-characterized receptors to identify common principles but also differences between receptor groups and classes. Novel aspects of signaling dynamics and protease-activated receptors will be included. Structural dynamics of GPCR activation and the modulation of receptor activation and signal selectivity, however, is the central question of all projects.
Overall, we aim to answer the following questions: How can we turn snapshots obtained by structural anal-yses in the first funding period into a consistent picture of the dynamic processes of GPCR activation and signaling? How do different types of ligands affect the dynamics and trafficking of GPCRs, and the interac-tion with different effector proteins? How is the interplay of extracellular and intracellular signaling and cou-pling mechanism orchestrated? How can we use the knowledge of the structural dynamics to obtain signaling protein profiles and to predict ligands and their activity?
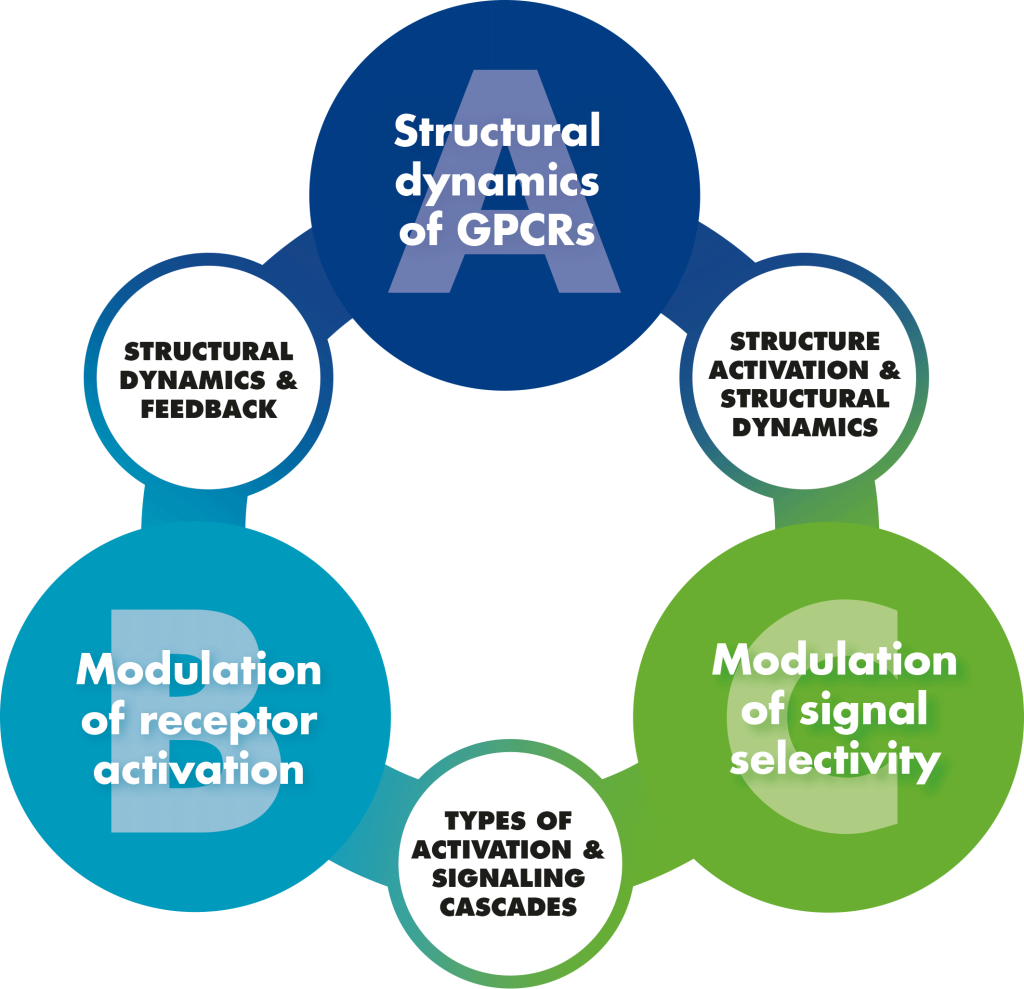
The SFB1423 is divided into four major groups:
- Identification of structural snapshots of peptide and adhesion GPCRs
- GPCR activity is modulated by distinct signals that lead to stabilization of active receptor conformations, which will be studied by
- Different conformations will lead to different signaling, e.g. G protein/arrestin bias, Gi/Gs dual coupling, cis/trans-signaling
- Peptide synthesis and membrane protein (GPCR) expression & Computational models of structure, dynamics and evolution of GPCRs

Scheerer
Structural elucidation of activity regulation at GPCRs involved in metabolism and immune responses

Huster, Schoeder
Investigation of the structure and dynamics of the ghrelin/GHS receptor complex

Elgeti, Schmidt
The role of conformational dynamics for Y receptor activation

Beck-Sickinger, Huster
Characterizing the molecular interaction between the Y receptor and arrestin

Scheerer, Schöneberg, Spahn
Structures of adhesion GPCR by cryo-electron microscopy

Langenhan, Sträter
Enzymology of autoproteolysis and signaling function of the GAIN domain in adhe-sion GPCRs

Meiler
Innovative artificial intelligence algorithms for modeling and docking of GPCRs

Beck-Sickinger
Molecular mechanisms of allosteric modulators at Y-receptors

Biebermann, Kühnen
Signaling dynamics of MC4R: development of advanced treatment strategies

Kaiser, Sinz
Transient contacts for ligand recognition and modulation of receptor activation by Y1R and Y2R N termini

Coin
Exploring topology and dynamics of molecular interactions at Y receptors in living cells via non-canonical amino acids

Liebscher
Signal integration and specification of adhesion GPCRs
Langenhan, Scholz
Dynamic modulation of adhesion GPCR function through complex formation

Hildebrand
Specificity and dynamics in signaling of peptide activated GPCRs

Annibale, Lohse (E)
Microscopic methods to study how GPCR dynamics and localization impact signaling specificity

Prömel, Schöneberg
The neglected receptor N terminus - signal filter, signal integration, trans-signaling of adhesion GPCR

Bock, Coin
Structural dynamics of allosteric coupling in GPCRs

Stäubert
Structure-, localization-, metabolism- and ligand-dependent signaling bias of metabolite-sensing GPCRs

Isermann, Künze
Allosteric regulation of PAR1 biased signaling by tethered peptide ligands and receptor oligomerization

Bock, Lohse
Mechanisms of receptor localization and localized signaling

Beck-Sickinger, Schöneberg
Central Administration

Huster
Integrated Research Training Group (IRTG)

Beck-Sickinger, Scheerer
Peptide synthesis and membrane protein (GPCR) expression

Hildebrand, Meiler, Stadler
Data management and computational models of structure and dynamics, and evolution of GPCRs