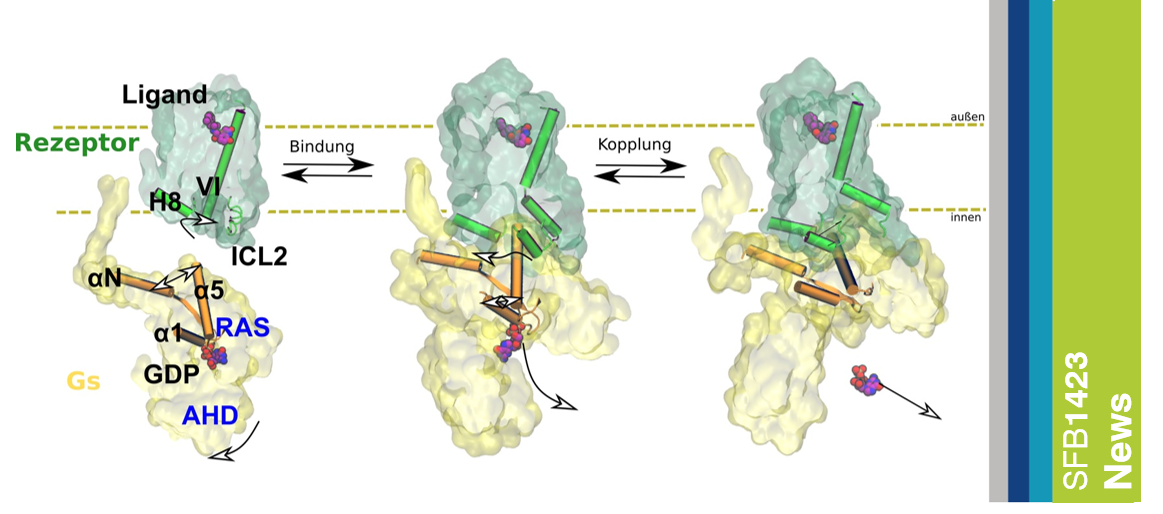
[Press release 2024/102 from 21/06/2024 by Tom Goetze] G protein-coupled receptors (GPCRs) are found throughout the human body and are


News & Events of the SFB1423
In addition to publishing current information on our research work, the SFB1423 also participates in numerous other formats such as conferences, lectures, workshops and exhibitions. On this page you will find a compilation of current news and events from our Collaborative Research Center.
Our Blog – be up to date
Insight into the molecular causes of
Thousands of people worldwide die every day from overdoses of opioids such as fentanyl. Drugs that act on the opioid
Speck and Heyder awarded by Ernst
In recognition of their outstanding work in the field of neuroendocrinology, David Speck and Dr. rer. nat. Nicolas Andreas Heyder
Season´s Greetings
Dear friends of the SFB1423,we thank you for the constructive and pleasant collaboration and we are looking forward to another
Meeting of the SFB1423 Project Leader
From May 11 – 13, 2022 the Project Leader and Members of the SFB1423 meets at the Hotel Kloster Nimbschen (Grimma).The
GPCRs – Universal genius among receptors
The Collaborative Research Center (CRC) 1423 "Structural Dynamics of GPCR Activation and Signal Transduction" under the leadership of biochemists Prof.
Events (upcoming)
We will have the Vanderbilt – Leipzig Symposium at Vanderbilt University, TN, USA from August 18 – 21, 2024.
If you are interesseted please send a E-Mail to .
Prof. Dr. Berend Isermann will give a short talk about his research group and his main research topics/goals.
“Biased signaling via PAR-receptors”
The next lecture will be on September 12. We would like to extend a warm welcome to Prof. David C. Martinelli, from the University of Connecticut, Farmington, USA (invited by Tobias Langenhan). The lecture will held in hybrid.
Lecture:
“The pleiotropic C1Q-like proteins regulate both synapses and oligodendrocyte maturation”
Zoom Link:
https://uni-leipzig.zoom.us/j/63731717253?pwd=dFcrdmxPNkFKd1FLa1JNM2JtY29NUT09
Venue:
Beckmann lecture hall
Faculty of Life Sciences
Brüderstraße 34, 04103 Leipzig
Meeting of the German Biophysics Society 2024
Please register here >>> https://dgfb2024.de/
As in previous years, the 27th STS Meeting will be organized as a Joint Meeting of the Signal Transduction Society with signaling study groups of the German Societies for Immunology (DGfI) and Cell Biology (DGZ), the Society for Biochemistry and Molecular Biology (GBM), the German Society for Pharmacology (DGP), and the Collaborative Research Center “Structural Dynamics of GPCR Activation and Signaling” (SFB1423).
More information and registration:
https://sigtrans.de/meeting-2024
>> abstract submission deadline: September 1, 2024 <<
We present our Collaborativ Research Center during the annual DIES ACADEMICUS 2024 at Leipzig University.
Twice a year all members of the SFB1423 meet for a retreat to discuss the progress of the project.
Vanderbilt-Leipzig MKF / SFB1423 Module
Computational Design and Directed Evolution of Therapeutic Peptides
Peptides as therapeutics are an emerging class of therapeutics modalities, due to their high binding affinities and specificities. Here we will discuss their characteristics as therapeutic class and give an overview on recent and future developments. Furthermore, we highlight strategies for identification and optimization of peptide therapeutics. We will also cover emerging technologies for structure-based computational design of peptides using Rosetta. Specifically, we will train students in the theoretical background of computational techniques for Peptide design and provide hands-on training with respect to engineering peptides consisting of genetically encoded but also non-canonical amino acids.
Date
December 16-20, 2024 / 2 – 6 PM
Venue
BBZ or S1/S2 Brüderstraße 34 and Virtually via Zoom.
Teacher
Prof. Allison Walker, PhD.
(Vanderbilt University, Department of Chemistry)
Prof. Annette Beck-Sickinger
(Leipzig University, Faculty for Life Science, Institute for Biochemistry)
Jun.-Prof. Dr. Christina Lamers, Jun.-Prof. Dr. Clara T. Schoeder, Prof. Dr. Leonard Kaysser, Dr. Georg Künze
(Leipzig University, Medical Faculty, Institute for Drug Discovery)
Prof. Dr. Jens Meiler
(Leipzig University, Medical Faculty, Institute for Drug Discovery & Vanderbilt University)
Registration
Students, postdocs, and faculty who wish to audit the class are welcome. Please send a note to Carie Fortenberry () and Anett Albrecht () as we have limited seating for the laboratory sections.
(posponed End 2024)
The next lecture will be on XYZ. We would like to extend a warm welcome to Prof. Dr. Hiroaki Suga, from the Tokyo, Japan (invited by Annette Beck-Sickinger).
Lecture:
“Pseudo-natural peptides, products and neobiologics for therapeutic applications”
The lecture will held in hybrid. Please register by sending an email to .
Events (past)
On behalf of Prof. Dr. Berend Isermann, we would like to cordially invite you to our next SFB1423 lecture.
Date & Venue:
June 27, 2024 at 5AM / Room „Justus von Liebig“
(Leipzig Medical Center, House 4, (Innere und Operative Medizin), Liebigstr. 20, 04103 Leipzig)
Speaker:
Dr. Silvio Antoniak
(University of North Carolina, USA)
Title:
“Protease-dependent GPCR activation in the cardiovascular system.“
If you want to join virtually, please use the Zoom link: https://uni-leipzig.zoom-x.de/j/65593302608?pwd=bm80STBsSlJVc2ZZR2FrRkdUZU5lUT09 (Code: 141325).
On behalf of Prof. Dr. Dr. Ines Liebscher, we would like to cordially invite you to our next SFB1423 lecture.
Date & Venue:
June 20, 2024 at 3AM / Beckmann Lecture hall
(Faculty of Life Sciences, Brüderstraße 34, 04103 Leipzig)
Speaker:
Prof. Dr. Jinpeng Sun
Prof. Dr. Zhao Yang
Prof. Dr. Pengju Zhang
Zili Liu
(Shandong University, Jinan, China)
Title:
“The discovery of the subfamily of steroid hormone membrane receptors and the perception of microenvironment by GPCR” (Sun/Yang)
“The roles and mechanisms of metabolite sensing GPCRs in cancer” (Zhang/Liu)
If you want to join virtually, please use the Zoom link: https://uni-leipzig.zoom-x.de/j/65593302608?pwd=bm80STBsSlJVc2ZZR2FrRkdUZU5lUT09 (Code: 141325).
The next lecture will be on May 21. We would like to extend a warm welcome to Prof. Dr. Johannes W. Hell, from the University of California, UC Davis, USA (invited by Andreas Bock). The lecture will held in hybrid.
Lecture:
“Role of regulation of the Ca2+ channel CaV1.2 by b-adrenergic signaling in attention”
Zoom Link:
https://uni-leipzig.zoom.us/j/63731717253?pwd=dFcrdmxPNkFKd1FLa1JNM2JtY29NUT09
Venue:
Beckmann lecture hall
Faculty of Life Sciences
Brüderstraße 34, 04103 Leipzig
Twice a year all members of the SFB1423 meet for a retreat to discuss the progress of the project.
The next lecture will be on February 7 from 11:30am – 1pm. We would like to extend a warm welcome to Dr. Obada Bahra from the Coskun Lab, Center of Membrane Biochemistry and Lipid Research, TU Dresden (invited by Tobias Langenhan). The lecture will held in presence.
Lecture:
“The lipid perspective of Akt signaling”
Venue:
Rudolf Schoenheimer Institute of Biochemistry
Johannisallee 30, 04103 Leipzig
Seminar Room D301/302 (3rd floor)
We are looking forward to your participation.
The 17th Leipzig Research Festival of Life Sciences takes place at the Leipzig University organized by the Faculty Of Medicine and the Faculty of Life Sciences. All young scientists can take the opportunity to present their latest research results in a public» Science Workshop «.
Awarding:
- Young Scientist Award
- Audience Award
Opening Abstract Submission: 16.10.2023
Deadline for Abstract: 04.12.2023
The next lecture will be on Monday, December 18. We would like to extend a warm welcome to Prof. Dr. Christa E. Müller, from the University of Bonn and Head of Department of Pharmaceutical & Medicinal Chemistry, Pharmaceutical Institute.
Lecture: “Structural Biology-Supported Drug Development for Purinergic Targets and Orphan G Protein-Coupled Receptors”
Twice a year all members of the SFB1423 meet for a retreat to discuss the progress of the project.
We present our Collaborativ Research Center during the annual DIES ACADEMICUS 2023 at Leipzig University.
Prof. Dr. Dr. Ines Liebscher will give a short talk about her research group and here main research topics/goals.
The next lecture will be on Tuesday, October 10. We would like to extend a warm welcome to Prof. Dr. Stefanie Schirmeier, from the Technische Universität Dresden, Faculty of Biology.
Lecture: “Metabolic flexibility of the nervous system”
The Lecture will be held at the RSI, Johannisalle 30, Seminar room – 3rd floor, 04103 Leipzig.
From September 24 to 26, Alexander von Humboldt Professor and SFB1423 project leader Jens Meiler and Junior Professor Clara T. Schoeder will host a lecture series on Artificial Intelligence in Drug Development at Leipzig University.
A key element of the summer schools will be presentation of their research by the doctoral researchers. These presentations will be given without the project leader of the project being present such that a good learning atmosphere will be created.
The next Summerschool will take place at the “Schlosshotel Schkopau” from Septermber 11 – 13, 2023.
Please contact Juliane Adler to get more details …
Prof. Dr. Daniel Huster will give a short talk about his research group and his main research topics/goals.
“Not a GPCR, but still interesting: the specificity of lipid interactions of the intramembrane protease GlpG”
Prof. Norbert Sträter will teach this module.
More information will follow soon…
The SFB1423 will be presented in differents events at the LONG NIGHT OF SCIENCE in Leipzig:
SFB1423 / GPCRs – Wir wollen wissen, wie sie funktionieren.
(SFB1423, BBZ, Deutscher Platz 5)
Wie Medikamente mit Rezeptoren tanzen
(Prof. Andreas Bock, Härtelstr. 16-18, SR429)
James Bond und die Physik
(Prof. Daniel Huster, Liebigstr. 13-27 GHS)
Going Deep: Künstliche Intelligenz in der Wirkstoffentwicklung
(Prof. Jens Meiler, Liebigstr. 13-27 KHS)
Pharmazeutische Wissenschaften – von der Idee zum Arzneimittel
(IfWE, BBZ, Deutscher Platz 5)
We cordially invite you to the Symposium of the Leipzig-Vanderbilt cooperation on June 9, 2023. International scientists of the Vanderbilt Diabetes Research and Training Center will talk about kidney-focused and diabetes-related research.
Ray Harris
Roy Zent
Ambra Pozzy
Ming-zhi
Venue: Small lecture hall, Talstrasse 33, ground floor
Time: 1 p.m.
MKF / SFB1423 Module
Integration of Experimental Data
with Artificial Intelligence to study
Membrane Proteins
Prof. Hassane Mchaourab
Vanderbilt University, Center for Structural Biology
Prof. Erkan Karakas
Vanderbilt University, Center for Structural Biology
Dr. Matthias Elgeti and Prof. Dr. Jens Meiler
Leipzig University, Faculty of Medicine
>> Click here to get the Program
Registration
Please send an Email to
.
More details will be announced shortly.
The next lecture will be on Thursday, May 25 from 4:30 – 6:30 PM. We would like to extend a warm welcome to Prof. Steffen Lindert, from the Columbia University, The Ohio State University, Dep. Of Chemistry and Biochemistry, USA with his lecture:
“COMPUTATIONAL PROTEIN STRUCTURE PREDICTION
FROM MASS SPECTROMETRY DATA”
You can join the lecture in person at the Charles Tanford Protein Centre, Halle (Saale), Seminar room E.04.0 or online via zoom (link will follow soon)
Twice a year all members of the SFB1423 meet for a retreat to discuss the progress of the project.
Prof. Dr. Tosten Schöneberg will give a short talk about his research group and his main research topics/goals.
„GPCR dysfunctions as causes of human pathologies“
The next lecture will be on Tuesday, April 4 from 2 – 3 PM. We would like to extend a warm welcome to Prof. Jonathan A. Javitch,, from the Columbia University, Department of Molecular Pharmacology and Therapeutics and Department of Psychiatry, USA with his lecture:
“My recent forays into adhesion GPCRs: what has stuck”
You can join the lecture in person at the Seminar Room (3rd floor), Rudolf Schönheimer Institute of Biochemistry, Leipzig.
Prof. Dr. Peter Hildebrand will give a short talk about his research group and his main research topics/goals.
„Structural mechanism of Gs protein activation by the prototypical beta-2 adrenergic receptor“
SFB1423/MKF Seminar:
Current Developments in Experimental and Computational engineering of Peptide Therapeutics
Peptides as therapeutics are an emerging class of therapeutics modalities, due to their high binding affinities and specificities. Here we will discuss their characteristics as therapeutic class and give an overview on recent and future developments. Furthermore, we highlight strategies for identification and optimization of peptide therapeutics. We will also cover emerging technologies for structure-based computational design of peptides using Rosetta. Specifically, we will train students in the theoretical background of computational techniques for Peptide design and provide hands-on training with respect to engineering peptides consisting of genetically encoded but also non-canonical amino acids.
Prof. Christina Lamers & Prof. Annette Beck-Sickinger (Leipzig University)
Prof. Allison Walker, Prof. Rocco Moretti and Prof. Benjamin P. Brown (Vanderbilt University)
Registration via email to Juliane Adler
The next lecture will be on Friday, January 13 from 9 – 10 AM. We would like to extend a warm welcome to Prof. Carsten Hoffmann from the Institute for Molecular Cell Biology of the University Hospital Jena with his lecture:
“GRK Knock-out cells reveal new insights into GPCR regulation”
You can join the lecture in person at the Institute of Biophysics, Lecture Hall (big), Härtelstraße 16/18 in Leipzig or you dial-in via Zoom link:
https://uni-leipzig.zoom.us/j/63261926503?pwd=cEwzSGVxYTJzY29NQUFscTd2OGtzZz09
On Thursday, January 12 from 9:15 – 10:15 AM Mr Fabian Pohl from the BBZ Leipzig, Sträter Lab will give a lecture at the Rudolf Schoenheimer Institute of Biochemistry on the topic:
“Structural Characterization of the Autoproteolysis and Dimerization Behavior of ADGRB2”.
You can join the lecture in person at the Rudolf Schoenheimer Institute of Biochemistry, Seminar Room D301/302 (3rd floor), Johannisallee 30 in Leipzig.